Enterococcal species have been introduced as the most important nosocomial pathogens in the recent years [1,2]. According to the surveillance system of hospital infections in the United States, enterococci are the fourth cause of hospital infections in the world [3]. Inspite of the fact that more than 20 enterococcal species have so far been identified, only E. faecalis and E. faecium species remain among the most important pathogens in human [4,5].
Since, enterococci are intrinsically resistant to most of common antibiotics and they have great potential to gain resistance to the antibiotics, anti-microbial treatment for enterococcal infections is complicated [6–8]. However, aminoglycosides and other antibiotics affecting the cell wall such as vancomycin and ampicillin are drugs of choice for the treatment of enterococcal infections. The recent studies showed that the antibiotic resistance patterns are rising among enterococci [7]. For example, more than 25% of enterococci represented resistance to aminoglycosides and more than 50% of E. faecium species indicated resistance to ampicillin [9].
Vancomycin Resistant Enterococci (VRE) was firstly reported in Europe in 1988 and then was noticed in USA in 1989 [10]. Today, VRE isolates have been known as one of the most important nosocomial infections and there are different reports about VRE prevalence all around the world.
It is well-known that resistance to vancomycin is due to the connective replacement of D-alanine in construction of peptide and glycan in the cell wall. In this process, final D alanine converts to the D lactate and vancomycin target site will disappear [11]. To date, nine different kinds of gene clusters including vanA, B, C, D, E, G, L, M, and vanN that mediate the resistance to vancomycin have been identified [12]. One of the most important genes is vanA. Phenotype of vanA is specified by high level resistance to the vancomycin and teicoplanin [8,13].
Because vancomycin is an effective treatment against enterococcal infections, resistance to this antibiotic among enterococci is of great importance [8]. Due to the presence of transferable resistant genes, VRE prevalence is increasing.
Phenotypic diagnostic methods such as disk diffusion and Minimal Inhibitory Concentration (MIC) as well as genotypic methods are currently used to identify VRE isolates [14]. However, it is necessary to find rapid, simple and alternative methods for detection of VRE isolates especially during prevalence of the isolates in the world [14,15].
Materials and Methods
In this cross-sectional study, a total of 200 isolates were collected from different clinical samples (urine, blood, wound, sputum and cervical secretion) between October 2012 to December 2012. The isolates were obtained from outpatients department who were admitted to four laboratories in the general and private hospitals in Tehran, Iran. Moreover, only patients who received no antibiotic therapy during the sample collection period or had no antibiotic treatment within a month prior to hospital admission were included in this study. The inclusion criteria were based on the standards defined by Center for Disease Control (Atlanta, GA, USA) [18]. The isolates were identified as Enterococci and identification of E. faecalis among the isolates was performed according to the standard microbial and biochemical methods such as catalase, Gram stain, growth in 6.5% sodium chloride medium, esculin hydrolysis and Pyrrolidonyl Arylamidase (PYR) test [19].
Disk Diffusion Test
The susceptibility of E. faecalis isolates to vancomycin was tested using the disk diffusion method. This method was performed according to the Clinical and Laboratory Standard Issue (CLSI) guidelines (2006) by using a disk containing 1 μg vancomycin (BBL, Sensi Disk, USA) to detect VRE isolates [17,20]. The procedure of this method has been previously described [21]. Zone size <14 mm was considered as resistant and zone size >17 mm was considered as sensitive [22].
Confirmation of the VRE isolates by PCR
DNA extraction of the E. faecalis isolates was performed by a DNA extraction kit (Roche, Germany). Briefly, the isolates were cultured in 5 ml of Luria Bertani (LB) medium. The bacterial pellets were then resuspended in lysis buffer containing lysozyme and incubated at 37°C for 30 min. After adding the precipitation solution, the solution was transferred to a spin column, washed with washing buffer, and DNA was eluted in the elution buffer.
Then, the vancomycin-resistant and vancomycin-sensitive E. faecalis isolates in disk diffusion method were tested to identify the presence or absence of the vanA gene using the PCR method. E. faecalis American Type Culture Collection (ATCC) 29212 strain was used as a positive control. The following primers were used for the amplification [23]:
Primer-F: TTAAAACCATTAGGCGATCG
Primer-R: CCCATTCCCATTGATGGATCCAT
PCR reaction was carried out in a final volume of 50 μl containing 2 μl of DNA template, 5 μl of 10x reaction buffer, 2 μl of dNTPs (10 mM), 2 μl of Mgcl2 (50 mM), 2 μl of each primer (10 pmol), and 1 U of Taq DNA polymerase (Fermentas; Vilnius, Lithuania). Then, the PCR products were electrophoresed on a 1% agarose gel to visualize under the Gel Doc UV light.
Fermentation of Turanose
Finally, all of the VRE and VSE isolates were considered for turanose fermentation. It was carried out according to the procedure previously described [21]. To find a considerable difference in metabolism of turanose among the confirmed VRE and VSE isolates, the growth of these isolates was assayed in the presence of different dilutions of turanose. Briefly, 150 μl of the nutrient broth (sugar free) medium containing 0.5%, 0.7% and 1% dilutions of turanose were inoculated to each well in 96-well microplates and suspensions of the cultured bacteria equal to 0.5 McFarland standards were added to the wells. Furthermore, the broth medium containing turanose without a microbial suspension was used as negative control for each dilution of turanose. Then, the inoculated microplates were incubated at 37°C for 24 hour and turbidity of the cultured isolates in the microplates was measured with ELISA reader at absorbance of 610 nm. The adsorption of the isolates was read 3 times to calculate the average of their adsorptions.
Statistical Analysis
The one-way (ANOVA), Student t-test and Tukey (HSD) test were used to compare the differences between the mean values of the groups using SPSS software. The p<0.05 of all results was considered as significant.
Results
Sample Identification
Out of 200 enterococcus isolates, 40 were identified as E. faecalis. According to the disk diffusion results, 20 E. faecalis isolates resistant to vancomycin (VRE) and 20 susceptible isolates (VSE) were identified.
Detection of VRE isolates by PCR
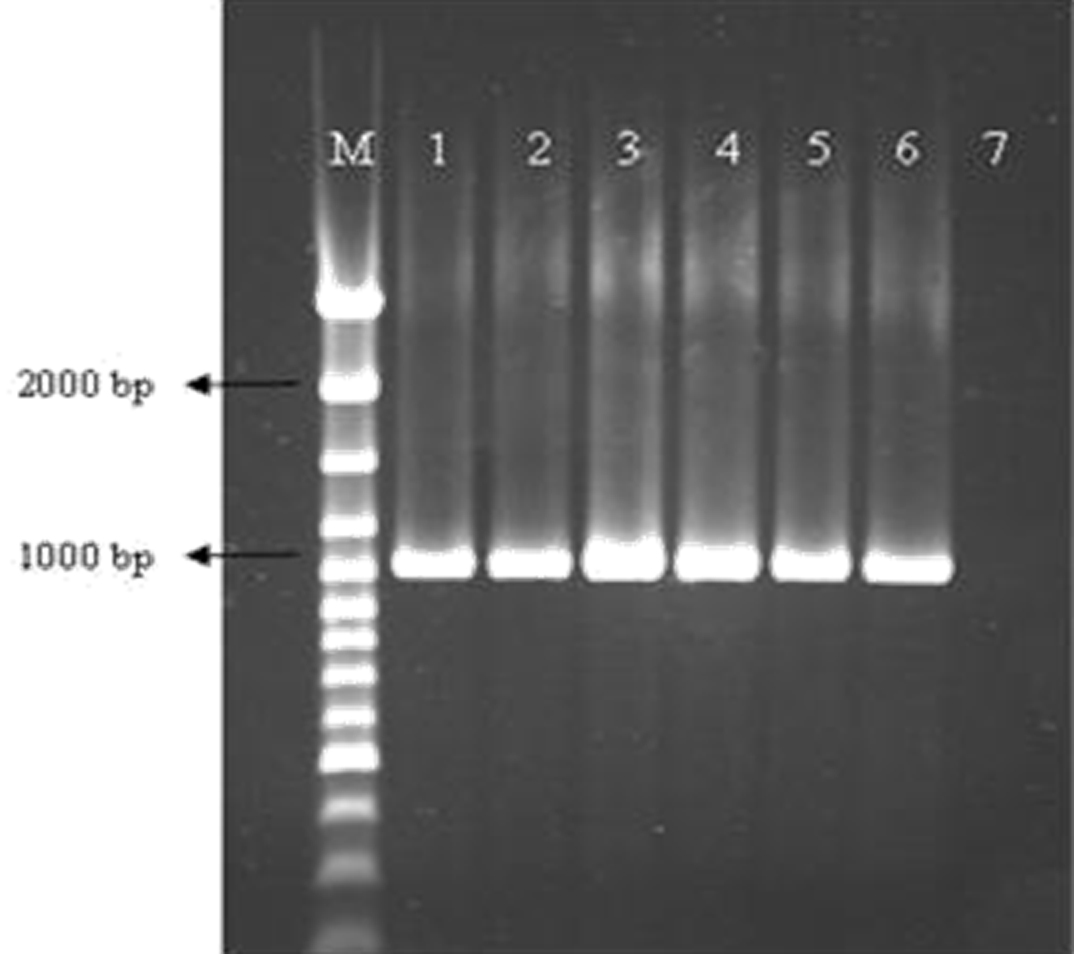
All of the VRE isolates were detected with the disk diffusion method contained the vanA gene in PCR reaction; whereas the vanA gene was not detected among the vancomycin-sensitive E. faecalis isolates. The results of PCR amplification of this gene indicated that the length of PCR product of vanA in VRE isolates was about 1030 bp [Table/Fig-1].
PCR product of the vanA gene in VRE isolates.
Line 1: positive control (E. faecalis ATCC 29212); Lines 2-6: vanA positive isolates; Line 7: Negative control; M: Molecular weight marker.
ATCC: American Type Culture Collection
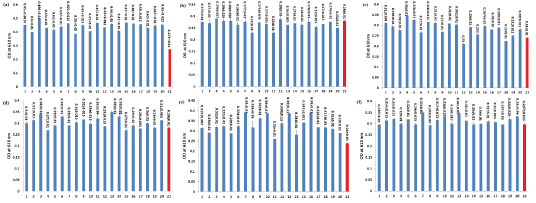
Turanose Fermentation in VRE and VSE isolates
We observed that all VRE isolates in 0.7% dilution of turanose had significantly higher absorbance than the negative control (p <0.05), whereas, none of the 20 VRE or VSE isolates in 0.5% and 1% dilutions of turanose had significantly more absorbance than the negative controls (p>0.05) [Table/Fig-2]. We also indicated that the absorbance of almost all of the VRE isolates (19 of 20 isolates) in 0.7% dilution of turanose was significantly higher than VSE isolates in dilutions 0.5%, 0.7% and 1% turanose (p<0.05), whereas the absorbance of VRE isolates in 1% and 0.5% dilutions of turanose was not significantly higher than VSE isolates in the turanose dilutions (p>0.05) [Table/Fig-2].
Fermentation of turanose by the VRE and VSE isolates in dilutions of 0.7%, 1% and 0.5%. The metabolism of turanose was evaluated at: a) dilution of 0.7% for VRE and; b) VSE,; c) dilution of 1% for VRE and; d) VSE, and; e) dilution of 0.5% for VRE and; f) VSE isolates. Numbers 1-20: the VRE or VSE isolates and number 21: negative control. The one-way ANOVA, Student’s t-test and Tukey HSD tests in the SPSS software were used for statistical analyses. There was a significant difference among VRE and VSE isolates in 0.7% dilution of turanose (p <0.05), whereas, there was no significant difference between VRE and VSE isolates in 1% and 0.5% dilutions of turanose (p >0.05). Bars represent mean±S.D. from three independent experiments.
Discussion
Enterococci are one of the most important causes of nosocomial and community-acquired infections in the world [24,25]. Furthermore, due to increased resistance and the ability to acquire resistance to new antibiotics quickly, it is more and more difficult to treat E.faecalis infections, especially with the emergence of vancomycin resistant E.faecalis (VRE) [8].
The variability in the prevalence of VRE from different reports was observed, such as 1% in Europe [26], 7% in the United State [27], 14.6% in India [22], 0.6% in China [9] and 9.5-16% in Iran [28].
Since enterococci have a remarkable potential to acquire and exchange antibiotic resistance genes, environmental enterococci are considered as a source of antimicrobial resistance genes that can transfer resistance to other pathogenic bacteria. In addition, there is an ongoing exchange among antibiotic resistance genes in clinical and environmental bacteria. This is a serious therapeutic challenge for treatment and public health [9,11].
Vancomycin is one of the most important alternative treatments against multidrug-resistant enterococcal infections [10,12]. High resistance to vancomycin among enterococci strains as well as the probability of transferring resistance genes to other Gram-positive bacteria such as Methicillin Resistant Staphylococcus aureus (MRSA) has made the Center of Disease Control (CDC) to offer guidance for prevention and control of VRE prevalence [22]. Thus, rapid detection of VRE by simple and low cost methods in clinical samples is very essential for the establishment of an antibiotic therapy system and prevention of the enterococcal infections.
Whereas, different methods have been used for diagnosis of the VRE isolates [14,29], there are some limitations in all of these methods. Molecular based methods, such as PCR are considered as a rapid method for detection of resistant isolates, but there are some VRE isolates which are not detectable with the PCR method [28]. For example, it has been reported that some resistant isolates had different genetic sequences and the designed primers failed to detect the VRE isolates in PCR reaction. Zhanel GG et al., showed that only 88.2% of VRE strains contained the vanA gene [30]. These differences in genetic sequences indicate that the necessary precaution should be noted in the application of genotypic methods in the detection of VRE isolates. Furthermore, the requirement for facilities and high costs does not permit many laboratories to use the PCR method for identification of VRE isolates. In addition to being time-consuming and expensive, some phenotypic methods such as disk diffusion also have difficulties in the diagnosis of VRE isolates [31]. In addition, MIC as another phenotypic method is not a precise and complete method for the detection of VRE isolates, because it is variable among E. faecalis isolates with vanA genotype (32-200 μg/m), making the detection of resistant strains difficult [14].
Thus, a diagnostic method identifying all VRE is currently missing and there is a need to evaluate other methods to develop a sensitive, specific and simple method for identification of the isolates [31].
The basis of phenotypic methods is the measurement of resulting products from biochemical reactions and these reactions are in fact an indirect assessment of bacterial genetics [29,32]. Since the other studies showed that genes responsible for resistance to antibiotics could make changes in the cell wall of Gram-positive bacteria, it was possible that there is a relationship between the cell wall changes by resistance genes and metabolism of sugars in these bacteria [33]. Therefore, in this study, we assumed that there is a relationship between metabolism of turanose with resistance to vancomycin in E. faecalis isolates. In our findings, the VRE isolates indicated statistical significance in 0.7% turanose over control group and all VSE isolates in 0.7%, 0.5% and 1% dilutions of turanose (p<0.05), whereas there was no significant difference between the metabolism of turanose in 1% and 0.5% dilutions in VRE isolates and VSE isolates (p>0.05).
The reasons for the low consumption of 0.5% and 1% dilutions of turanose in VRE isolates are unknown and need to further studies are required to explore the reasons behind it, but we have several possibilities for it: 1) These dilutions of turanose may influence the phenotypic expression of resistance gene to vancomycin in VRE isolates; and 2) These dilutions of turanose may alter the cell wall permeability especially the importer pumps of turanose that resulted in the low penetration of turanose into the cytoplasm of the VRE isolates.
Interestingly in other study, Raeisi J et al., reported a significant relationship between the consumption of turanose at 0.7% dilution and resistance to methicillin in Staphylococcus aureus isolated from clinical samples [21]. In other study, Morgan JW et al., demonstrated that all novobiocin-resistant Staphylococcus strains fermented turanose, whereas novobiocin-susceptible Staphylococci strains failed to ferment turanose [34].
Limitation
The sample size was small and further studies with larger sample size are required to present this method as a reliable detection method in the future.
Conclusion
Because of the high prevalence of E. faecalis infections and due to the increased resistance among the isolates and other bacteria, performance of such studies that can find a simple method to diagnose resistant strains can be highly valuable. In this study, we suggest the turanose metabolism at 0.7% dilution as a simple and novel phenotypic method that can be used for the diagnosis of VRE isolates. This was a preliminary study and it is obvious that a bigger population of the isolates has to be analysed in order to introduce this method as a reliable method for detection of VRE isolates. It is difficult to compare the efficacy of this method with other routine detection methods of VRE isolates, however if we want to compare the results of turanose fermentation in 0.7% dilution with PCR and disk diffusion methods in this study, all of the methods almost had 100% efficacy in detection of VRE isolates. Finally, further studies should be done on the metabolism of other sugars than turanose for better differentiation of VRE from VSE isolates.