Atopic dermatitis is a recurrent chronic condition that generally affect young children [1]. The aetiology of this disease is multifactorial involving an intricate play of the host genetic composition and environmental factors [2].
On understanding the immunological aspect of the AD, it has been shown that cytokine imbalance may be responsible for the initiation of inflammation. Cytokines like IL-4, IL-5, IL-10 have been implicated in the production of Ig E, by activation of B cell population and role of IL-13 resulting in a class switch of Ig E in allergic diseases [3]. Malassezia species-specific IgG and IgM antibodies can be detected in healthy individuals as well as atopic dermatitis patients, however these antibodies do not contribute in protection against the disease as known with other allergic diseases [4]. However, healthy individuals are usually not sensitized to Malassezia spp., as compared to AD patients [5].
A SNP in a cytokine gene may also affect the production of cytokines. Genetic predisposition studies on various diseases have shown relationship between cytokine gene polymorphism and host susceptibility to disease [6–11].
Materials and Methods
The study was conducted for the period of 12 months (January 2012 to January 2013) including enrolment, analysis and compilation of data.
The sample size included 38 patients [14] of consecutive untreated clinically diagnosed cases of AD attending Outpatient Department of Dermatology and Venereology in a tertiary Care hospital (University College of Medical Sciences and Guru Tegh Bahadur Hospital, Delhi, India), irrespective of age and sex.
The disease was characterised by severely itchy, red, dry, and crusted skin with the distribution varying with age. There is usually a history of other atopic diseases. Exclusion criteria were history of light therapy, antimycotic treatment in last two months, topical antifungals within one month, topical corticosteroids within one week, immunocompromised or suffering from severe systemic illness.
Skin scrapings were collected by using a blunt sterile scalpel from different body sites (shoulder, neck, back, chest, arms, cheek, nasolabial folds, and forehead) [15]. Thirty eight, age matched healthy unrelated individuals without a clinical history of any skin disease were considered as healthy controls. A clearance from ethical committee was obtained as per the institutional guidelines. Informed consent was obtained from the patients. Every patient was given the right to opt out from the study at any stage of project.
The skin scrapings were subjected to conventional laboratory methods for the identification of Malassezia yeast, such as KOH examination and culture on non selective media (Sabouraud dextrose agar with Chloramphenicol and Cyclohexamide), and selective media (Modified Dixon’s Agar with Chloramphenicol and Cyclohexamide). The media was incubated aerobically at 250C. The colonies appeared within 8-10 days but incubation was continued for four weeks before being discarded as sterile. Stock cultures were maintained at -200C in sterile for further use.
Speciation of the Malassezia
Speciation was done using the phenotypic method, which included morphological characteristics (macroscopic morphology and microscopic morphology), and physiological methods (Urease test, Catalase test, β-Glucosidase test, Tween Assimilation test and Growth at 370C and 400C) [16].
A 3 ml venous blood sample in EDTA vial was also collected aseptically from all patients for DNA extraction and subsequent study for SNP. The blood samples were stored at 40C, till further use.
Genomic DNA extraction
Genomic DNA was extracted from blood samples of all study subjects for determining three SNPs (IL10-1082A/G; IL10-819/ 592C/T; IFN-γ+874A/T) in two cytokine genes IL 10 & IFNγ through ARMS-PCR using sequence specific primers [17].
Genomic DNA was extracted from EDTA anticoagulated peripheral blood using HiPurATM blood genomic DNA extraction kit (Himedia Laboratories) following manufacturer’s instructions.
A 200 μl of blood sample was collected in a 2.0 ml collection tube, 20 μl of the reconstituted proteinase K solution (20 mg/ml) was added. The sample was vortexed for 10-15 seconds to ensure thorough mixing. To obtain RNA free genomic DNA, 20 μl of RNase A solution (20 mg/ml) was added and mixture vortexed again for 10-15 seconds. The sample was then incubated for two minutes at room temperature (15-250C). Following this, 200 μl of the lysis solution (C1) was added to the sample and vortexed thoroughly for a few seconds to obtain a homogenous mixture. The sample was incubated at 550C for 10 minutes in a water bath. The lysate for binding to the spin column, was prepared as follows; 200 μl ethanol (96-100%) was added to the lysate obtained from the above step and mixed thoroughly by gentle pipetting. Lysate was transferred into the spin column provided with the kit and centrifuged at 10,000 rpm for one minute. The flow-through liquid was discarded and the column placed in a new 2.0 ml collection tube. A 500 μl of diluted pre wash solution was added to the column and centrifuged at 10,000 rpm for one minute. After discarding the flow-through liquid, 500 μl of diluted wash solution was added to the column and centrifuged at 13,000-16,000 rpm for three minutes to dry the column. Flow- through liquid was discarded and a dry spin was given at the same speed to remove the residual ethanol, if any. The column was placed in a new 2.0 ml collection tube and 200 μl elution buffer was added without spilling to the sides. The column was incubated at room temperature for five minutes for high yield of DNA and then centrifuged at 10,000 rpm for one minute to elute the DNA. The samples were stored at -200C until used. DNA samples were subjected to specific PCR reaction in cytokine genotyping.
Primers for SNP Polymorphism [
18,
19]
The following were the sequence:
IL-10 -1082G/A
Common Primer: 5’ – CAGTGCCAACTGAGAATTTGG – 3’
G allele: 5’ – CTACTAAGGCTTCTTTGGGAG – 3’
A allele: 5’ – ACTACTAAGGCTTCTTTGGGAA – 3’
IL-10 -819C/T / -592C/A
Common Primer : 5’ – AGGATGTGTTCCAGGCTCCT – 3’
C allele : 5’ – CCCTTGTACAGGTGATGTAAC – 3’
T allele : 5’ – ACCCTTGTACAGGTGATGTAAT – 3’
IFN-γ +874T/A
Common Primer: 5’ – TCAACAAAGCTGATACTCCA – 3’
A allele: 5’ – TTCTTACAACACAAAATCAAATCA – 3’
T allele: 5’ – TTCTTACAACACAAAATCAAATCT – 3’
B-globulin (Internal control):
Forward: 5’ – ACACAACTGTGTTCACTAGC – 3’
Reverse: 5’ – CAACTTCATCCACGTTCACC – 3’
Cytokine genotyping was carried out from genomic DNA by ARMS-PCR with sequence specific primers (Sigma Aldrich, Bangalore, India) [18,19]. Three SNPs (IL10-1082A/G; IL10-819/592C/T; IFNγ +874A/T) in two cytokine genes were assessed in all the patients and healthy controls. The PCR products were loaded onto a one percent agarose gel in a specific order for electrophoresis and run at 150 volts for 20-25 minutes for separating the DNA. After electrophoresis, ethidium bromide stained gel was photographed and interpreted for specific amplification patterns. Presence of a control band in each lane was ascertained (β globulin, 100 bp). Wells identifying the IL10-1082 and IL10-819/592 cytokines contained a band of 258bp and 233bp respectively [18]. Wells identifying the IFNγ +874 cytokines contained a band of 261bp [20].
Statistical Analysis
Comparison was obtained using Chi-Square test or Fisher’s-Exact test (wherever applicable). Odds ratio and 95 percent Confidence Interval was calculated and p<0.05 was taken as significant.
Cytokine polymorphisms and genotype frequencies were evaluated by gene counts. The observed and expected genotype frequencies data was analysed using Chi-Square test. As multiple comparisons were made, Bonferroni’s correction was applied to significant p values (p<0.02) that were multiplied for the number of genotypes detected [21]. But, as p<0.05 is also considered as statistically significant in small study group, our discussion included all variables considering p<0.05 as significant.
Results
In AD group, Malassezia yeasts were cultured in 24 out of 38 samples and thus the identification rate was 63.1 percent. Four Malassezia species (Malassezia globosa, Malassezia sympodialis, Malassezia furfur, Malassezia restricta), were identified as the common species in AD group patients. The species isolated from healthy control were Malassezia globosa, Malassezia sympodialis, and Malassezia furfur. The overall identification rate was 52.6 percent (20/38) [Table/Fig-1].
Malassezia yeasts between patients and healthy adults.
| Species | AD 63.1% (24/38) | Healthy 52.6% (20/38) |
|---|
| Malassezia globosa | 34.2% (13/38) | 39.5% (15/38) |
| Malassezia sympodialis | 15.8% (6/38) | 10.5% (4/38) |
| Malassezia furfur | 2.6% (1/38) | 2.6% (1/38) |
| Malassezia restricta | 10.5% (4/38) | 0% (0/38) |
Cytokine gene polymorphism in Atopic Dermatitis
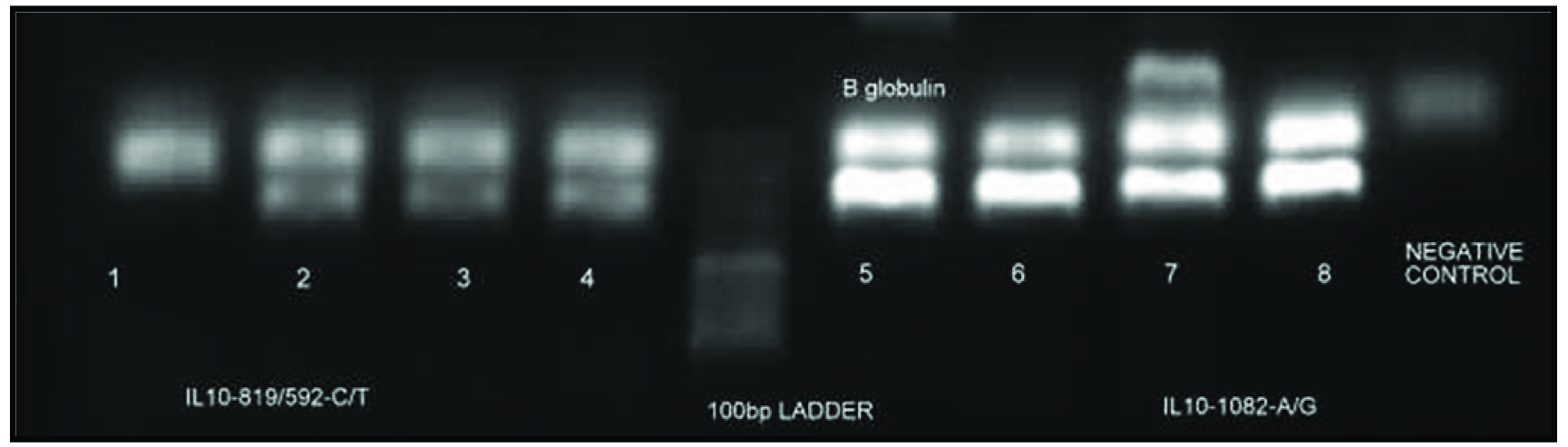
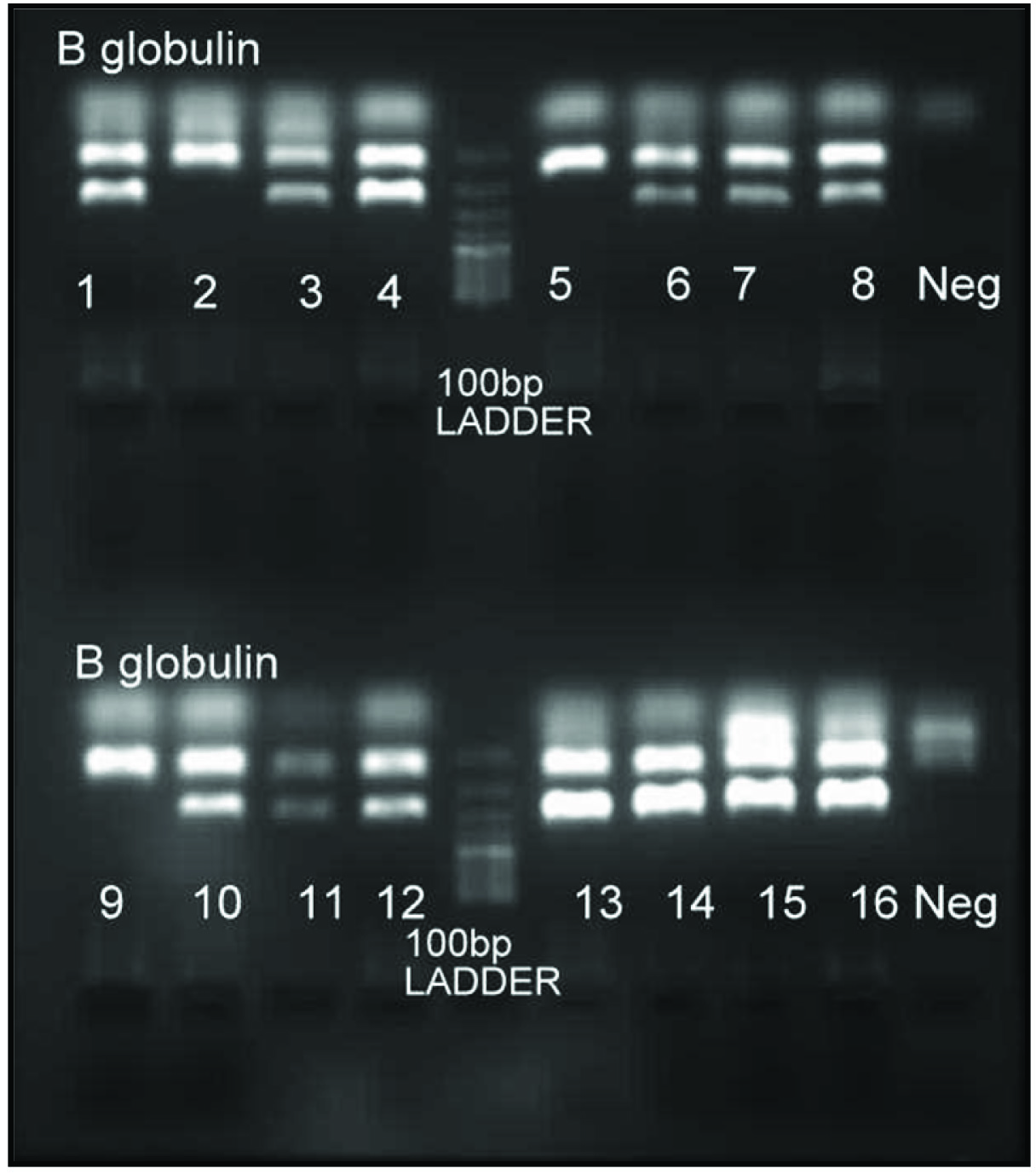
The PCR products were analysed by gel electrophoresis using ethidium bromide stained gel [Table/Fig-2,3].
ARMS-PCR analysis for genotyping IL10-819/592C/T and IL10-1082A/G polymorphism {(1, 2)-IL10-819/592TT; (3, 4)-IL10-819/592CT; (5, 6;7, 8)- IL10-1082AG}.
ARMS-PCR analysis for genotyping IFNg+874 T/A polymorphism {(1,2)-IFNg+874TT; (3,4;7,8;11,12;13,14;15,16)-IFNg+874AT; (5,6;9,10)- IFNg+ 874AA}.
Patients with AD (n=38; 25 males, 13 females), and healthy controls (HC) (n=38; 21 males and 17 females), the genotype distribution of all cytokine SNPs followed the Hardy-Weinberg distribution and showed no deviation from the HWE (p>0.001).
In our study [Table/Fig-4], significant differences in allele and genotype distribution was observed in IL10–1082G/A, IL10-819/592C/T and IFNγ +874T/A gene polymorphisms.
Allele and genotype frequencies of cytokine polymorphisms in Atopic Dermatitis patients and healthy controls.
| Cytokine Polymorphism | | | AD (N=38) | HC (N=38) | p-value | Odds ratio | 95% CI |
|---|
| IL10-1082 | Alleles | A | 61(80.3%) | 45(59.2%) | 0.005*# | 2.801 | 1.354-5.795 |
| G | 15(19.7%) | 31(40.8%) | 0.005*# | 0.357 | 0.173-0.738 |
| Genotypes | AA | 23(60.5%) | 7(18.4%) | 0.000*# | 6.790 | 2.384-19.343 |
| AG | 15(39.5%) | 31(81.6%) | 0.000*# | 0.147 | 0.052-0.419 |
| GG | 0(0.00) | 0(0.00) | - | - | - |
| IL10-819/592 | Alleles | C | 34(44.7%) | 46(60.5%) | 0.51 | 0.528 | 0.277-1.006 |
| T | 42(55.3%) | 30(39.5%) | 0.051 | 1.894 | 0.994-3.610 |
| Genotypes | CC | 4(10.5%) | 9(23.7%) | 0.128 | 0.379 | 0.106-1.360 |
| CT | 26(68.4%) | 28(73.7%) | 0.613 | 0.774 | 0.286-2.092 |
| TT | 8(21.1%) | 1(2.6%) | 0.028* | 9.867 | 1.168-83.351 |
| IFN+874 | Alleles | A | 24(31.6%) | 40(52.6%) | 0.009*# | 0.415 | 0.215-0.804 |
| T | 52(68.4%) | 36(47.4%) | 0.009*# | 2.407 | 1.243-4.662 |
| Genotypes | AA | 2(5.3%) | 12(31.6%) | 0.003*# | 0.120 | 0.025-0.584 |
| AT | 20(52.6%) | 16(42.1%) | 0.358 | 1.528 | 0.618-3.779 |
| TT | 16(42.1%) | 10(26.3%) | 0.147 | 2.036 | 0.774-5.358 |
*Significant according to (p<0.05)
# Significant according to Bonferroni’s correction (p<0.02), AD-Atopic Dermatitis, HC- Healthy Control
The distribution of IFNγ+874T/A (p=0.009) and IL10-1082A/G (p=0.005) alleles were significantly different between patients and HCs. AD patients were more likely to carry the IL10–1082 G allele (p=0.005) and it was significantly associated with the disease (OR=0.357, 95% CI 0.173-0.738). IFNγ+874 A allele was significantly associated with AD (OR=0.415, 95% CI 0.215-0.804). No statistically significant differences in allele distributions were found between the patients with AD and controls for IL10-819/592.
The distribution of IL10-1082 AA and AG genotype was significantly different between patients and controls (p<0.001). IL10-1082 AG genotype was significantly associated with AD (OR=0.147, 95% CI 0.052-0.419). In AD patients, the IL10-819/592 TT genotype frequency was higher in patients in comparison to HC (21.1% vs 2.65, p=0.028). Similarly, the distribution of IFNγ+874 AA genotype was significantly associated with AD (OR=0.120, 95% CI 0.025-0.584).
Discussion
AD is a frequent chronic relapsing inflammatory skin disorder. It is characterised by intensely itchy skin eczema and is frequently associated with allergic rhino-conjunctivitis and allergic asthma. The prevalence of AD increased during the last few decades, affecting 15-30% children and upto 10% adults [1,22]. Malassezia yeasts are normal flora found on the skin of 75~98% of healthy persons [23]. Malassezia yeast has been found to constitute 80% of the total skin fungal population in both humans and animals [24].
Despite its diagnosis and chronic nature, the aetiopathogenesis is not fully understood. The dysregulation between host genetics and several environmental factors are responsible for the development of AD [2].
It is well known that the skin of AD patients has an impaired skin barrier function [25] and an altered immune system depicted by high IL- 4, IL-10 and IL-13 levels compared to healthy individuals [26,27]. These cytokines reduce the production of the antimicrobial peptides LL-37, human beta defensin (hBD)-2, and hBD-3 in keratinocytes [27–29], which are important components of the skin’s innate immune system in the defence against microorganisms. This suggest that the impaired skin barrier function along with the altered skin immune system may play intertwining roles and contribute to colonization and growth of microorganisms on the skin of AD [25,29]. Role of microorganism in the exacerbation of AD has been studied in the past with patients suffering showing sensitivity to Malassezia allergens [30,31]. Such patients may benefit from an antifungal therapy.
Several studies investigated the epidemiology of Malassezia spp. in healthy and AD patients by culture and molecular methods obtaining variable results presumably because of differing methodologies. The apparent variability in the data may be explained by differences in race, region, ethnicity, or, geographical occurrence of Malassezia.
Malassezia yeast in AD act as an allergenic aggravating factor rather than as infectious agents. 30%–80% of adult AD patients have a positive skin prick test with Malassezia spp. extract [32]. The pH of the skin surface in patients with AD is higher than that of normal healthy skin suggesting a host microbe interaction where Malassezia will release more allergens into the skin environment thus contributing to the inflammation [13].
In this study, Malassezia globosa was the most commonly isolated species in both patient and healthy control followed by Malassezia sympodialis, Malassezia furfur and Malassezia restricta. Other studies report either Malassezia globosa or Malassezia sympodialis as the most common species in AD patients [33]. In a study of Nakabayashi A et al., the most common species in lesional skin of AD Japanese patients was M. furfur (21%) [34]. Other culture independent studies detected M. globosa and M. restricta as the predominant species in AD [35]. Canadian and Korean study had M. sympodialis as the predominant species in AD patients, with a detection rate of 51.5% and 16.3% respectively [36,37].
It has also been observed that probably due to its slow growth, Malassezia restricta, was overgrown by other isolates and related species. Host genetic factors play a major role in determining the differential susceptibility to infectious diseases of humans. Cytokine production can influence both disease manifestation and severity; therefore, cytokine gene polymorphism could determine the susceptibility of the host to disease [38]. The production or function of cytokines is, atleast partially, regulated by polymorphisms in their gene sequences [38]. IL10 shifts Th1/Th2 balance by down regulating the Th1 response and suppression of proinflammatory cytokines IFNγ, secretion. IL10 is a Th2 antiflammatory cytokine that can suppress the immune response and inter individual variation in IL10 production are genetically determined by polymorphism within the IL10 promoter region-1082 G/A, -819 C/T and -592 C/A. The polymorphism at -819 C/T and -592 C/A are in linkage disequilibrium with each other [18].
IFNγ is a Th1 proinflammatory cytokine that can augment the immune response. The functional SNP accusation +874 T/A is located at the 5’ end of a CA repeat at the first intron of human IFNγ gene. The T allele correlates with high IFNγ expression [19].
The homozygous TT genotype is associated with the ability to produce high levels of IFNγ, the heterozygous TA genotype with intermediate levels, and the homozygous AA genotype with lower levels [14].
In our study, polymorphism in the gene IFNγ at position +874 T/A in the first intron was identified in AD patients. IFNγ+874 A allele was significantly associated with AD. IFNγ+874 AA genotype frequency was found to be higher in AD patients than in controls. This finding suggests that AD patients may produce a lower IFNγ. We postulated that the time of production and concentration of proinflammatory cytokines during the inflammation process may be critical but due to allelic polymorphisms, a dampened T cell response was observed.
In AD patients, the IL10-819/592 TT genotype frequency was higher in patients in comparison to HC.
The distribution of IL10-1082 A/G alleles was significantly different between AD patients and HCs. AD patients were more likely to carry the IL10–1082 G allele and it was significantly associated with this disease. IL10-1082 AG genotype was significantly associated with AD.
IL10 driven Ig E antibody production as an agent of underlying atopy also need to be evaluated [39]. In recent years, much research has been done on the involvement of multiple susceptibility genes working in concert with Treg cells to produce abnormal phenotype in several skin diseases. IL-10 polymorphism may suggest that the imbalance from the deficiency in the suppression by the Regulatory T cells (Treg cells) or a strong activation signal that supersede the regulatory mechanism, can result in the predominance of IL-10 production as suggested by ourfinding in AD (IL10-1082 AG). However, this has to be documented in terms of numerical and functional differences of Treg population in AD from that of HCs.
Other cytokines like IL-2 levels can be detected to understand its role as an immunomodulator and recovery of Treg population. It is worth noting that the sequence of four Malassezia allergens, namely Mala s6, 10, 11, 13 reveals high homology to human endogenous proteins [33].
The demonstrated cross reactivity of Mala s6 and Mala s13 with human cyclophilin and thiorexidin respectively suggest that autoimmunity due to molecular mimicry may have an important role in the pathogenesis of AD, a further research may contribute to its association [33].
Limitation
The limitation of the study was use of phenotypic methods for culture and not molecular approach for isolation. Many species especially the slow growing ones like M. restricta are difficult to grow. Also, there was a lack of comparison of Cytokine gene polymorphism data with the serum cytokine levels in the study group for better understanding of the genetic susceptibility of the host to Malassezia infections. Data on population genetics require inclusion of larger number of subject to evaluate the probability or the frequency of occurrence of the genotype but due to time constraints of a year for completion of this project along with inclusion of other disease like Pityriasis versicolor and Seborrhoeic dermatitis rendered it difficult to increase the study group size.
Conclusion
In conclusion, isolation rate from culture studies was higher in cases as compared to HC suggesting higher prevalence on skin of Malassezia yeast in atopic patients. Thus, elaborating the need to do further studies focussing on species specific characterisation with identification of virulence traits. Also, the cytokine gene assessment result along with the presence of clinical outcome in this study helped to build a hypothesis of association between susceptibility to a disease in presence of the right environmental triggers in a suitable host. This study can be used further to direct the research for predicting the probability of disease manifestation from a commensal organism in genetically susceptible host.