Chronic Periodontitis, a progressive inflammatory disease of multifactorial nature characterized by tooth-supporting tissues destruction, is one of the most prevalent diseases in the world [1]. It results from an imbalance between the load of periodontopathic bacteria in the subgingival microenvironment and the immunological potential of the host, which can be altered by numerous risk factors, including demographic, behavioural, environmental, and systemic aspects [2,3], as well as genetic susceptibility determined by genotype [2–6].
In this sense, it has been reported that several genetic polymorphisms which encode mediators of bone homeostasis have been found to be associated with periodontitis susceptibility [5–7]. Taking into account that vitamin D and its receptor VDR play special roles in various physiological processes including bone and calcium metabolism, cellular growth and differentiation, and immunity [8], mutations in functionally critical areas of VDR gene might influence the development of periodontitis not only through their immunomodulatory effects, but also via their effects on bone mineral density [9,10]. The gene encoding the VDR is located on chromosome 12q13.11 (OMIM 601769), harbours eight exons that are invariably translated, and six that are alternatively spliced. The VDR gene exhibits several SNPs, located in both the coding and the non-coding portions of the gene [11]. Whilst both rs7975232 (ApaI) and rs1544410 (BsmI) SNPs are found in the region of the gene from intron 8 to the 3’ untranslated region, a silent mutation within codon 352 of the exon 9 creates the rs731236 (TaqI) polymorphic site [9]. In addition, the genetic variation rs2228570 (FokI), located in the exon 2 at the 5’ portion [12], has been regarded as a start codon polymorphism [13].
Materials and Methods
Study Design
This cross-sectional, observational, analytic study was conducted in accordance with the ethical guidelines of the Helsinki Declaration and was evaluated and approved by the Ethics Committee for Human Studies of the Faculty of Dentistry, University of Antioquia in Medellín, Colombia. The sample size was calculated on the basis of a previous study regarding the association of VDR gene polymorphisms with periodontal disease [14]. It was increased by 20% to maintain the estimates at an optimal level of precision (5%) against the potential effect of sample size reduction due to exclusions and dropouts. Thus, the theoretical sample size for clinical screening was set to 150 individuals (distributed between two groups) to determine significant differences in outcomes at the 95% confidence level, with an alpha value = 0.05 and 80 % power. However, every effort was made to recruit the maximum number of participants, so that the study sample included a total of 160 participants from the population of individuals that sought treatment and/or consultation at the Graduate Periodontics Clinics. Afterwards, the study power was analyzed using a website genetic power calculator tool (http://pngu.mgh.harvard.edu/~purcell/gpc/cc2.html) with the following parameters: prevalence of CP in Colombia (0.50) [23], equal frequencies of marker alleles (0.20) with disequilibrium coefficient (D’) = 1, genotype relative risk (2.0), a dominant model of inheritance, and control/case ratio (0.45).
Volunteer Recruitment
In order to be considered for inclusion in the study, the volunteers had to be of Colombian ancestry. Informed consents were signed by all volunteers prior to their enrolment into the study. Complete demographic and medical data (including gender, age, medications, systemic health, and smoking habit) were obtained from all participants. Individuals were considered as non-smokers if they had never smoked, or had quit smoking at least five years before to the date of examination [24]. All recruitments underwent both a clinical assessment to rule-out any pathological condition of the oral mucosa and a clinical periodontal examination. A full-mouth periodontal examination was performed by a single trained and calibrated observer using a manual periodontal probe (PCP UNC 15, Hu-Friedy, Chicago, Illinois, USA) for determining the Probing Depth (PD) and Clinical Attachment Level (CAL) at six sites around each tooth, except for third molars. These measurements were rounded off to the nearest millimetre and were used to estimate both the extent and severity of periodontitis based on the percentage of tooth sites having PD ≥4 mm along with CAL ≥2 mm (extent) and the average value of attachment loss of the diseased sites (severity), according to well-defined criteria [25].
Inclusion and Exclusion Criteria
The study population comprised 110 untreated CP patients and 50 periodontally HC. The CP group comprised patients having at least 20 remaining teeth, periodontal disease as evidenced by at least four tooth sites with PD ≥4 mm and CAL ≥2 mm [26], evidence of alveolar crestal bone loss ≥2 mm evaluated on digital periapical radiographs as the distance from the Cemento-Enamel Junction (CEJ) to the most apical extension of the bony defect at two sites per tooth (mesial and distal) under standard conditions [27], as well as the presence of consistent amounts of bacterial/mineralized plaque deposits regarding to the severity of periodontal breakdown. Healthy controls comprised individuals with no history of periodontal disease, pocket depth ≤3 mm, and no clinical gingival inflammation (no more than 10% of sites with bleeding on probing and absence of gingival redness/oedema), but could have attachment loss or gingival recession due to mechanical trauma. Exclusion criteria applied were presence of diseases affecting the structural integrity of the oral hard and soft tissues, except dental caries and periodontal disease (e.g., lichen planus, pemphigus psoriasis, aphthous stomatitis); ongoing orthodontic therapy; chronic medical conditions affecting the host’s periodontal status and bone metabolism (e.g., osteoporosis, connective tissue diseases, endocrine diseases, immunological disorders); or that would require pre-medication for monitoring or treatment procedures (e.g., heart conditions, joint replacements, corticosteroid or antiresorptive therapy); a course of anti-inflammatory, mouthrinses, or antibacterial agents for the preceding three months; and periodontal treatment within the last six months. Moreover, diseased subjects were excluded if they met the any of criteria for aggressive periodontitis (e.g., rapid and severe attachment loss/bone destruction, relative low levels of biofilm and calculus regarding to the severity of periodontal destruction) [25].
Saliva Sample Collection and DNA Extraction
Prior initial periodontal treatment period, 5 ml of unstimulated whole saliva was collected from each individual by spitting into a 50 ml sterile plastic centrifuge tube (Greiner Bio-one®, Frickenhausen, Germany) in the morning just before breakfast consumption and any dental hygiene procedure. After collection, whole saliva was clarified by centrifugation for five minute at 800 x g (IEC® Centra CL2 Centrifuge, Thermo Electron Corporation, Milford, Massachusetts, USA). The obtained pellet was dispersed using a vortex (IKA® Vibrofix, Staufen, Germany) for 15 seconds and 200 μl were used for DNA extraction using the QIAamp® DNA mini kit (Qiagen Sciences®, Germantown, Maryland, USA). DNA was stored frozen at -20°C until use.
Detection of VDR-ApaI, -BsmI, -FokI and -TaqI Polymorphic Sites
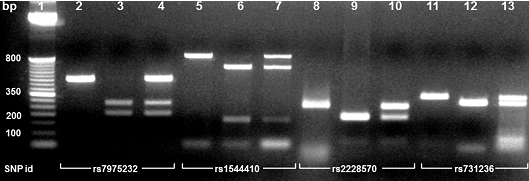
Polymorphic sites were determined by the Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) method. Specific primers used for determining the ApaI [16], BsmI [16], FokI [28], and TaqI [15] polymorphic sites in the VDR gene are listed in [Table/Fig-1]. PCR amplification was carried out in a 96-well thermal cycler (Mastercycler® gradient, Eppendorf®, Hamburg, Germany). Samples were initially denatured at 95°C for 10 minutes, followed by 35 cycles of 95°C for 45 seconds, 65-66°C for one minute, 72°C for one minute, and a final extension step of 72°C for two minutes. The annealing temperatures were determined by gradient PCR, selecting by agarose gel electrophoresis the best resolution in the expected fragment. PCR products were run in 2% agarose gel electrophoresis, stained with 0.5 mg/ml ethidium bromide and visualized in an ultraviolet transilluminator (FOTO/Analyst® Apprentice, Fotodyne Inc., Hartland, WI, USA). The size of the expected fragments before enzymatic digestion were 490 bp for rs7975232 (ApaI), 825 bp for rs1544410 (BsmI), 265 bp for rs2228570 (FokI), and 340 bp for rs731236 (TaqI). As molecular size marker, a 50-to-800 bp in multiples of 50 bp and additional fragment at 2652 bp DNA ladder was used. For negative control, DNA sample was replaced by sterile water.
Description of the VDR gene SNPs under study according to National Centre for Biotechnology Information (NCBI). wt, homozygous wild type; ht, heterozygous mutated; mut, homozygous mutated; bp, base pairs; SNPs, single nucleotide polymorphisms.
| SNP id | Forward primer/reverse primer | Restriction enzyme | Localization | Reference sequence: position | Base change | Genotype1 | Restriction enzyme digest fragment size (bp2) |
|---|
| rs7975232 | 5’-CAG AGC ATG GAC AGG GAG CAA G-3’5’-CAC TTC GAG CAC AAG GGG CGT TAG-3’ | ApaI | Intron 8 | NG_008731.1:g.64978 | G>T | TT (mut)GT (ht)GG (wt) | 490490, 280, 210280, 210 |
| rs1544410 | 5’-CAA CCA AGA CTA CAA GTA CCG CGT CAT GA-3’5’-AAC CAG CGG GAA GAG GTC AAG GG-3’ | Mva1269I (BsmI) | Intron 8 | NG_008731.1:g.63980 | G>A | AA (mut)GA (ht)GG (wt) | 825825, 650, 175650, 175 |
| rs2228570 | 5’-AGC TGG CCC TGG CAC TGA CTC TGC TCT-3’5’-ATG GAA ACA CCT TGC TTC TTC TCC CTC-3’ | FokI | Exon 2 | NG_008731.1:g.30920 | T>C | CC (mut)TC (ht)TT (wt) | 265265, 169, 96169, 96 |
| rs731236 | 5’-CAG AGC ATG GAC AGG GAG CAA G-3’5’-GGA TGT ACG TCT GCA GTG TG-3’ | TaqI | Exon 9 | NG_008731.1:g.65058 | T>C | CC (mut)TC (ht)TT (wt) | 260, 80340, 260, 80340 |
Restriction Endonuclease Digestion
PCR products from VDR gene polymorphic sites were digested with the respective restriction enzymes (Thermo Scientific®, Lafayette, CO, USA) in a volume of 20 μl containing 2 μl of 10X restriction buffer, 0.1 μl of restriction enzyme and 10 μl of PCR product. For BsmI site, an isoschizomer (Mva1269I) was used. To test the optimal activity, an analytical scale restriction enzyme digest was performed on 0.5-1.0 μg of lambda DNA (Fermentas®, Glen Burnie, MD, USA) for ApaI (1 recognition site) and ϕX174 RF1 DNA (Fermentas®) for Mva1269I (BsmI), FokI, and TaqI restriction enzymes (4, 8, and 10 recognition sites respectively) as digestion control. Reactions were performed in a digital dry bath (Accublock®, Labnet International, Edison, NJ, USA) and the incubation conditions were: ApaI, 30°C for one hour; Mva 1269I (BsmI), 37°C for two hour; FokI, 37°C for 10 minutes; and TaqI, 65°C for two hours. Lambda and ϕX174 RF1 DNA controls were used also as a control of digestion during all assays with participant’s samples. Fragments were separated in agarose 2% stained with ethidium bromide. [Table/Fig-1] shows the complete description of the VDR gene SNPs genotyping method. Substitution naming followed the nomenclature suggested by the HUGO Gene Nomenclature Committee [29].
Statistical Analysis
All data collected were analyzed using the statistical package IBM® SPSS® 23.0 (Chicago, Illinois, USA). Intra-observer reproducibility for clinical measures of PD and CAL, as well as for PCR assays was determined through double evaluations for each specific test performed by the same observer with ten participants selected randomly using a computer-generated randomization code (Epidat 4.0®, PAHO/WHO, Washington, DC, USA). The interval between tests one and two was seven days. For comparisons, the reliability between the two series of data was assessed by using the Cohen’s kappa statistic (κ) to evaluate the reproducibility of categorical variables and the Intraclass Correlation Coefficient (ICC) was selected when variables were quantitative.
All grouped data were tested for normality using the Kolmogorov-Smirnov test. Because the data were normally distributed, statistical tests were performed using parametric methods. Deviation from Hardy-Weinberg Equilibrium (HWE) was evaluated by goodness-of-fit by comparing the genotype frequencies with those expected on the basis of the observed alleles using χ2 critical value test with 1 degree of freedom. The odds associated with individual genotypes and alleles were calculated as the Odd’s Ratio (OR) with their 95% CI. Also, an analysis of the interaction between genetic findings and those significant demographic factors was performed for all SNPs. LD and haplotype frequencies distribution resulting from these four SNPs were analyzed using Haploview 4.2 software (MIT/Harvard Broad Institute, Cambridge, Massachusetts, USA). LD was measured based upon calculating disequilibrium coefficients (D’) and plots were drawn with the same application. Haplotypes were constructed with frequency threshold for rare haplotypes of <5%. All tests were two-sided and statistical significance was assumed at a p-value <0.05.
Results
Post-Hoc Genetic Power Calculation and Reproducibility of Measurements
Genetic power calculations suggested that the sample of 110 cases and 50 controls would provide >86% power to reject the null hypothesis of no association at p<0.05 when a genotypic relative risk was ≥2.0. Intra-observer reproducibility was excellent for both PD (ICC = 0.915, p=0.019) and CAL (ICC = 0.921, p=0.017) scores in each series of measures recorded per patient by the same examiner. Equally, intra-observer reproducibility was excellent for PCR assays (κ=1.00).
Demographic and Clinical Profile of the Study Population
Demographic and clinical characteristics of participants recruited for this study are summarized in [Table/Fig-2]. As it can be seen, not only the number of females, but also data regarding mean age and smoking habit were significantly increased (p<0.05, χ2 and unpaired t-test) in the CP group in comparison with healthy controls, so that these three variables met the criteria to be considered confounders for the association between genetic findings and CP. As expected, mean values of both PD and CAL scores in HC were significantly lower than those of CP patients (p<0.001, unpaired t-test). CP group included individuals with different extent of the periodontal damage which varied from a relatively localized to a generalized form of the disease. In the same way, participants included a representative sample of individuals presenting different degrees of the disease which varied from a slight to a severe form of periodontal breakdown.
Summary of demographic and clinical characteristics of the study participants according to diagnosis category.
| Characteristics | Clinical groups | p-value |
|---|
| Healthy controls(n = 50) | Chronic periodontitis(n = 110) |
|---|
| Gender* | Male | 23 (14.40) | 33 (20.60) | 0.049¶ |
| Female | 27 (16.90) | 77 (48.10) |
| Age (years)† | 33.46±11.57 | 47.09±10.24 | <0.001# |
| Smoking habit* | Non-smokers | 44 (27.50) | 72 (45.00) | 0.003¶ |
| Smokers | 6 (3.75) | 38 (23.75) |
| PD score (mm)†‡ | 2.02±0.26 | 2.84±0.57 | <0.001# |
| CAL score (mm)†‡ | 0.76±0.69 | 3.08±1.10 | <0.001# |
| Extent of periodontitis*§ | Localized (≤30%) | -- | 82 (74.50) | |
| Generalized (>30%) | 28 (25.50) |
| Severity of periodontitis*|| | Slight (1-2 mm) | -- | 8 (7.27) | |
| Moderate (3-4 mm) | 39 (35.46) |
| Severe (≥5 mm) | 63 (57.27) |
* Values are given as n (%) of subjects
† Values are given as mean±SD
‡ Data based on measurements obtained from the entire dentition
§ Percentage of periodontal pockets ≥ 4mm deep and attachment loss ≥ 2 mm
|| Data based on average value of CAL of the affected tooth sites
¶ Two-sided Pearson’s chi-square test (χ2)
# Two-sided unpaired t-test
Identification of VDR Genotypes and Analysis for Association with CP
Different banding patterns detected in agarose gel electrophoresis allowed the identification of genotypes for each SNP [Table/Fig-3]. The distribution of genotypes and alleles of VDR gene SNPs in periodontally HC and CP patients is depicted in [Table/Fig-4]. Both groups were in Hardy-Weinberg equilibrium with non-significant χ2 values comparing the observed and expected genotype frequencies (p>0.05, χ2 < 3.841). In relation to rs7975232 polymorphism, it was noteworthy that the allele T was carried by 85.0% (136 out of 160) of the participants; of these, 29.4% (47 out of 160) were homozygous. Likewise, regarding rs1544410 polymorphism, allele A was carried by 57.5% (92 out of 160) of the participants; being 15.0% (24 out of 160) homozygous. Alternatively, allele C of rs2228570 polymorphism was carried by 87.5% (140 out of 160) of the participants; of which, 36.3% (58 out of 160) were homozygous. Finally, allele C of rs731236 polymorphism was carried by 53.8% (86 out of 160) of the participants; of which only 9.4% (15 out of 160) were homozygous. Despite of aforementioned observations, the results showed that there was no association neither between the different genotypes nor allele frequencies and CP (all p>0.05, χ2). Similarly, no significant differences in extent or severity amongst genotype or allele groups were observed (all p>0.05, data not shown). Even so, logistic regression analysis revealed that synergistic interactions between each SNP and age were significantly associated with the disease status [Table/Fig-5]. All other interactions were not statistically significant.
Ethidium bromide-2% stained agarose gel showing PCR-RFLP salivary analysis of the VDR gene. Lane 1, 50 bp DNA ladder. Lane 2: homozygotic PCR product which is not susceptible to ApaI digestion (TT); Lane 3, homozygotic digestion (GG); Lane 4, heterozygotic digestion (GT). Lane 5, homozygotic PCR product which is not susceptible to BsmI (Mva1269I) digestion (AA); Lane 6, homozygotic digestion (GG); Lane 7; heterozygotic digestion (GA). Lane 8, homozygotic PCR product which is not susceptible to FokI digestion (CC); lane 9, homozygotic digestion (TT); Lane 10, heterozygotic digestion (TC). Lane 11, homozygotic PCR product which is not susceptible to TaqI digestion (TT); Lane 12, homozygotic digestion (CC); Lane 13, heterozygotic digestion (TC).
Genotype distributions and allele frequencies of VDR gene polymorphisms under study by clinical group.
| SNP id (restriction enzyme) | Clinical groups* | OR (95% CI)† | p-value‡ |
|---|
| Healthy controls(n = 50) | Chronic periodontitis(n = 110) |
|---|
| rs7975232 (ApaI) |
| Genotype | GG | 5 (10.0) | 19 (17.3) | Referent | 0.110 |
| GT | 25 (50.0) | 64 (58.2) | 1.48 (0.50 -4.40) |
| TT | 20 (40.0) | 27 (24.5) | 2.82 (0.90 - 8.92) |
| Allele frequency | G | 35 (35.0) | 102 (46.4) | Referent | 0.057 |
| T | 65 (65.0) | 118 (53.6) | 1.60 (0.98 – 2.61) |
| Hardy-Weinberg equilibrium χ2 critical value | 0.978 | 2.892 | | |
| χ2 test p value | 0.322 | 0.089 | | |
| rs1544410 (BsmI) |
| Genotype | GG | 18 (36.0) | 50 (45.5) | Referent | 0.504 |
| GA | 23 (46.0) | 45 (40.9) | 1.42 (0.68 - 2.97) |
| AA | 9 (18.0) | 15 (13.6) | 1.67 (0.62 - 4.47) |
| Allele frequency | G | 59 (59.0) | 145 (65.9) | Referent | 0.233 |
| A | 41 (41.0) | 75 (34.1) | 1.34 (0.82 – 2.18) |
| Hardy-Weinberg equilibrium χ2 critical value | 0.242 | 0.798 | | |
| χ2 test p value | 0.622 | 0.371 | | |
| rs2228570 (FokI) |
| Genotype | TT | 6 (12.0) | 14 (12.7) | Referent | 0.706 |
| TC | 28 (56.0) | 54 (49.1) | 1.21 (0.42 - 3.49) |
| CC | 16 (32.0) | 42 (38.2) | 0.89 (0.29 – 2.71) |
| Allele frequency | T | 40 (40.0) | 82 (37.3) | Referent | 0.642 |
| C | 60 (60.0) | 138 (62.7) | 0.89 (0.54 – 1.44) |
| Hardy-Weinberg equilibrium χ2 critical value | 2.777 | 0.252 | | |
| χ2 test p value | 0.095 | 0.615 | | |
| rs731236 (TaqI) |
| Genotype | TT | 21 (42.0) | 53 (48.2) | Referent | 0.767 |
| TC | 24 (48.0) | 47 (42.7) | 0.77 (0.38 – 1.57) |
| CC | 5 (10.0) | 10 (9.1) | 1.48 (0.24 – 2.59) |
| Allele frequency | T | 66 (66.0) | 153 (69.5) | Referent | 0.527 |
| C | 34 (34.0) | 67 (30.5) | 0.85 (0.51 – 1.40) |
| Hardy-Weinberg equilibrium χ2 critical value | 0.483 | 0.006 | | |
| χ2 test p value | 0.486 | 0.935 | | |
* Values are given as n (%) of individuals within diagnosis
† Odds ratio (95% confidence interval)
‡ Two-sided Pearson’s chi-square test (χ2)
Interactive effects amongst VDR gene polymorphisms and significant demographic variables regarding chronic periodontitis.
| Biological interactions | OR (95% CI)* | p-value† |
|---|
| rs7975232 (ApaI)GenderAgeSmoking habit | 1.30 (0.56 -3.06)1.12 (1.07 - 1.17)0.36 (0.12 - 1.07) | 0.541<0.0010.070 |
| rs1544410 (BsmI) GenderAgeSmoking habit | 1.37 (0.59 - 3.17)1.12 (1.07 - 1.16)0.39 (0.13 - 1.12) | 0.460<0.0010.081 |
| rs2228570 (FokI)GenderAgeSmoking habit | 1.40 (0.61 - 3.23)1.11 (1.07 - 1.18)0.39 (0.13 - 1.14) | 0.429<0.0010.085 |
| rs731236 (TaqI)GenderAgeSmoking habit | 1.39 (0.60 - 3.22)1.11 (1.07 - 1.16)0.38 (0.14 - 1.15) | 0.436<0.0010.081 |
* Odds ratio (95% confidence interval)
† Wald test
Furthermore, to control and minimize any source of variability attributable to demographic-related variations, both genotype and allele frequencies were analyzed after an individual’s stratification. Accordingly, no statistical differences could be noted amongst gender, age stratum (≤30 years vs >30 years), and smoking habit subgroups with respect to genetic variants under study (all p>0.05, χ2; data not shown), thus reinforcing the optimal comparability of the data amongst participants.
Linkage Disequilibrium and Haplotype Analyses
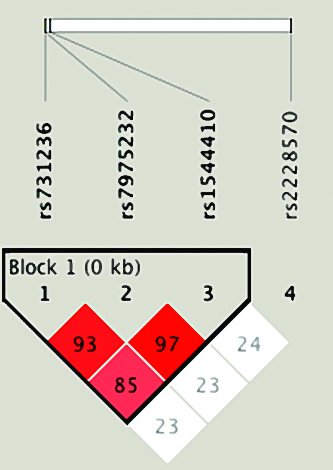
[Table/Fig-6] shows the LD patterns of four SNPs on VDR gene. As it is evident from this figure, three SNPs, which are physically very proximal, are in high LD. It is noticeable that the only LD block was formed by alleles at rs731236, rs7975232, and rs1544410 SNPs. It is also clear that SNP rs2228570 does not constitute a LD block in the sample studied, which makes sense given the physical distance from the other SNPs. The inferred haplotype distributions for healthy controls and CP patients are listed in [Table/Fig-7]. For statistical advantage, six haplotypes with a frequency of <5% were excluded from further analysis. The remaining seven haplotypes accounted for 96.55% of estimated haplotypes in this Colombian population. Nevertheless, the haplotype association analysis revealed that none of the haplotypes formed was significantly related to the risk of CP in the current study.
Linkage Disequilibrium (LD) block of the VDR gene markers typed in this study. The figure shows that rs731236, rs7975232, and rs1544410 SNPs belong to the same block of high LD.
Distribution of VDR haplotypes by clinical group and their association with the risk of chronic periodontitis.
| Haplotype* | Clinical groups† | OR (95%CI)§ | p-value|| |
|---|
| Healthy controls‡(n = 97) | Chronic periodontitis‡(n = 206) |
|---|
| GGTT | 22 (22.68) | 59 (28.64) | Referent | |
| GGCT | 13 (13.40) | 40 (19.42) | 0.69 (0.21 - 2.25) | 0.540 |
| TACC | 26 (26.80) | 50 (24.27) | 0.56 (0.23 - 1.35) | 0.200 |
| TGCT | 14 (14.43) | 31 (15.05) | 0.58 (0.20 - 1.69) | 0.320 |
| TATC | 7 (7.22) | 9 (4.37) | 0.36 (0.11 - 1.25) | 0.110 |
| TACT | 6 (6.19) | 10 (4.85) | 0.61 (0.19 - 1.98) | 0.410 |
| TGTT | 9 (9.28) | 7 (3.40) | 0.17 (0.03 - 0.95) | 0.045 |
* The order of the polymorphism is as follows: rs7975232, rs1544410, rs2228570, and rs731236
† Values are given as n (%) of haplotypes within diagnosis
‡ Six haplotypes with a total frequency of <5% were excluded from analysis: 3 HC and 14 CP cases
§ Odds ratio (95% confidence interval)
|| Two-sided Pearson’s chi-square test (χ2)
Discussion
In view of each individual has a singular dose-dependent response to the bacterial load that defines the susceptibility to periodontitis [17], over recent decades, immunogenetic studies have provided insight into the individual differences in the pathogenesis of CP and several recent publications have shown that susceptibility to dysbiotic microbial communities with potential for destructive inflammation is based on host genetic factors that may predispose to or protect from disease. Hence, dysbiosis alone may not necessarily precipitate CP, but it could induce disease in the context of other risk variables, such as host genotype, ageing, and/or behavioural habits such as smoking [6,30,31]. Although the genetic basis for CP has been thoroughly acknowledged [31,32], there is still controversy concerning the role of specific genes, such as VDR, that encodes the VDR, which is involved in a variety of biologic processes, including bone metabolism, modulation of the immune response, as well as regulation of cell proliferation and differentiation [13]. Likewise, VDR is essential for the maintenance bone’s structural integrity and exerts a profound effect not only on potent osteoclastogenic cytokines, including Interleukin (IL)-1 and -6, and tumour necrosis factor alpha [33], but also on lymphoproliferative response and macrophage mRNA synthesis and phagocytosis [34]. Owing to these facts, the current study investigated whether SNPs in VDR gene are linked with the periodontal clinical status in a group of Colombian individuals.
The search for genetic contributions to complex conditions such as CP is challenging due to different factors such as the genetic heterogeneity, incomplete penetrance, as well as the gene-gene and gene-environmental interactions [35]. This investigation constitutes an explorative approach assessing changes in the detection frequency of VDR SNPs from human genomic DNA obtained from saliva of the study participants in order to identify potential associations with CP in Colombian population. The foremost findings reported here was that although all genotype variants were detected both in the patients and control saliva samples, none of SNPs were associated with CP and did not have influence on the different degrees of extent/severity of the disease. Although the present findings parallel, at least partially, those reported by others using venous blood [9,16] or oral mucosa samples [36], opposed results have also been described [1,10,15,17,19,20]. Not with standing, it is important to point out that the observed discrepancies may be mainly due to the distinct populations can exhibit different genotype/allele frequencies which may be attributed essentially to racial and geographical factors and that may complicate interpretation of the results of genetic studies [37]. The Colombian population consists of a large ethnic mixture of individuals of different ethnic background [38]. This circumstance makes very difficult to match the ethnicity of patients and healthy controls and may have accounted for some confounding effects of undetectable population stratification.
It has been acknowledged that SNPs represent natural sequence variants, which may occur with more than one form, having a frequency greater than 1% in a human population [39]. Nevertheless, both genotype and allele frequencies may vary between different ethnic groups, being more or less homogenous between populations sharing common ancestries or underlying different phylogeographical origins [40]. In the present study, allele T carriage rate of rs7975232 (ApaI) SNP was 85.0%, while other studies show the following carriage rates: 80.9% [41], 66.1%, and 58% [16]. For rs1544410 (BsmI) SNP, allele A carriage rate was 57.5%, while for other populations was 40.1% to 67% [16,41,42]. Alternatively, allele C of rs2228570 (FokI) SNP was carried by 87.5% of the participants, nearly analogous to what was observed for other populations: 88.3% [41] and 76.3% [42]. Similarly, for the rs731236 (TaqI) SNP the frequency of allele C carriers (53.8%) was comparable with that of the different populations: 63.4% [41], 51.7% [16], and 41.2% [42]. Consequently, taking into account that the high frequency observed in the mutated alleles of each SNP is comparable to that of other populations, but does not constitute a risk for CP in the present study population, it could be assumed that the functionality of these SNPs and their causative role might include possible interactions with other factors such as demographical, environmental, and bacterial pathogens, to modulate susceptibility to disease [35].
According to the former, while this study failed to confirm an independent association amongst the four VDR SNPs with disease status, a synergistic interaction effect was significantly detected between each SNP regarding the individual’s age. This result appears to indicate that ageing cumulative characteristics of periodontal damage are significant modifying factors for the effect of VDR SNPs on CP. Although nothing is known about this biological interaction on which a base of comparison, could be established in agreement to the former, several publications [43–49] have been acknowledged synergistic risk patterns between ageing and genetic factors in CP. Ageing has been associated with histological and clinical changes in oral tissues, including epithelium thinning and diminished keratinization due to alterations occurring in gingival epithelial and connective tissue components [43]. Likewise, it is known that ageing alters the maintenance of bone remodelling and metabolism, presumably leading to an increased resorption of bone matrix [49,50]. In addition, it has been recognized that both downregulation of adaptive immune system and upregulation of the innate immune system pathways of older individuals may result in chronic inflammation [51]. Altogether, these data suggest that accumulation of molecular, cellular, structural, and functional noxious challenges to the integrity of periodontal tissues over time along side with an individual genetic predisposition increase susceptibility to subgingival bacterial colonization and might result in increased risk of the elderly to CP [43,49]. Confirmation of this finding and additional research into the underlying mechanisms responsible for these interactions besides the inclusion of different genetic variants are required to dissect their functional consequences and to evaluate their potential causative roles in CP.
Since rs731236, rs7975232, and rs1544410 SNPs were in strong LD in the present study, it would be possible to assume that these SNPs could not contribute to the CP susceptibility independently, nor VDR haplotypes inferred were associated with the risk of periodontal disease. These findings concur with previous reports where extensive LD at the BsmI, ApaI, and TaqI polymorphic sites was found [15,16]. Moreover, it has been postulated not only that the BsmI, Apal, and Taql polymorphic sites do not appear to alter VDR gene expression or VDR function, but also that pathological associations with these polymorphisms are most likely due to LD with other functional variations within the VDR gene or with another closely linked gene or genes [12]. Also, although the rs2228570 (Fokl) polymorphism remains as a potentially functional variant, it does not appear to be in LD with the Bsml, Apal, or Taql polymorphic sites in some populations [12,20,42], thus suggesting that there might be additional functional polymorphisms in the VDR gene that remain to be characterized. To finalize, certain limitations including the cross-sectional study design, the relatively small sample size, the variability of patterns of disease amongst the study subjects, and the large ethnic mixture of individuals, were associated with this study. Therefore, longitudinal studies with larger population samples would have greater statistical power and precision to evaluate the role of VDR SNPs in the pathogenesis of CP.
Conclusion
Although the present results do not support that VDR SNPs could be identified as independent risk predictor variable for CP in the present population, synergistic biological interactive effects of all these SNPs related to age might play a significant role in the pathogenic pathways of CP. Taking into account that the information concerning gene polymorphisms as susceptibility factors of periodontal disease may be useful in assessing disease progression and in treatment planning, the data presented suggest that VDR SNPs might represent a modifier of age of onset rather than a susceptibility factor for patients with CP.