Introduction
Breast cancer is the most common cancer in women and the Erythroblastosis Oncogene B(ErbB) receptor family holds crucial role in its pathogenesis. Human Epidermal Growth Factor Receptor 3 (HER-3) gene over expression in breast tissue has been associated with aggressive clinical behaviour and bad prognosis.
Aim
To evaluate HER-3 mRNA expression level as a prognostic marker for breast cancer and to correlate its level with other established prognostic parameters.
Materials and Methods
This study was carried out on specimens of 100 cases that were divided into 40 patients presented with fibroadenoma and 60 patients presented with Invasive Ductal Carcinoma (IDC) not otherwise specified and underwent modified radical mastectomy. All specimens were investigated for HER-2/neu, ER and PR expression by Immunohistochemistry (IHC) and quantitative assay of HER-3 mRNA expression using real time PCR technique.
Results
There was a significant high HER3 mRNA level in carcinoma cases compared to fibroadenoma. In malignant cases, HER3 mRNA level was significantly associated with advanced T stage, advanced N stage, number of positive lymph nodes, large tumour size and cases associated with an adjacent in situ component. Moreover, HER-3 mRNA level was of highest values in Her-2/neu positive group followed by triple negative cases with the lowest level in luminal group (p<0.05).
Conclusion
HER-3 gene is upregulated in IDC especially those carrying poor prognostic features. HER-3 mRNA level may identify a subset of patients with a poor prognosis, and who could undergo further evaluation for the efficacy of HER3 targeted anticancer therapy.
Introduction
Breast cancer is the most common cancer diagnosed in women and the most common cause of female cancer death in both developed and developing countries [1]. In Egypt, breast cancer is still the most common cancer in women with a lower incidence rate than that of USA, however, demographic studies demonstrated that breast cancer will be a greater public health concern in Egypt in the future [2].
Type 1 Growth Factor Receptor (T1GFR) family consists of four known members: HER-1 (Human Epidermal Growth Factor Receptor -1) [also known as Epidermal Growth Factor Receptor, (EGFR), and c-erb B-1], HER-2 (c-erb B-2), HER-3 (c-erb B-3), and HER-4 (c-erb B-4) [3]. The T1GFRs are trans membrane receptors with an extracellular ligand-binding domain, an α-helical trans-membrane segment, intracellular tyrosine kinase domain, that exist as monomers in their inactivate form, and a C-terminal phosphorylation tail [4]. Upon ligand binding the T1GFRs form homodimers or heterodimers (with themselves or other family members) and initiate a signaling cascade through activation of its tyrosine kinase, leading to cellular proliferation and transformation [5].
HER-3 was first identified in 1989 by Kraus et al., and is encoded by the ERBB3 gene [6]. The human ERBB3 gene is located on the long arm of chromosome 12 (12q13). It is encoded by 23,651 base pairs and translates into 1342 amino acids [7]. The ErbB2-ErbB3 heterodimers are considered the most active of the possible ErbB dimers, in part because ErbB2 is the preferred dimerization partner of all the ErbB family members, and ErbB3 is the preferred partner of ErbB2 [8].
ErB receptor family consists of HER-1, HER-2 and HER-3, where the latter cannot form homodimer and has a weak intra-cellular tyrosine kinase activity [9]. C-terminal region of HER-3 can bind the SH2 domain of Phosphatidylinositol, 3 Kinase/Akt PI3K by six consensus phosphotyrosine sites, implicating its crucial role in the activation of the PI3K/Akt pathway. It seems clear that activation of PI3K/Akt signaling by HER-3 is able to overcome HER-targeted inhibition. HER-3 is capable of restoring its signaling activity despite inhibition of other ErbB kinases due to the redistribution of signaling functions to different ErbB family members [10]. This character of HER-3 is not noticed in EGFR, HER-2, and HER-4. Expression and translocation of HER-3 from the nucleus to the membrane was reported to be responsible for resistance to EGFR or HER-2 targeted therapy [11].
Activation of HER3 leads to HER3 phosphorylation and subsequent activation of signaling pathways, such as phosphatidylinositol 3-kinase (PI3K)/Akt and RAF/MEK/Extracellular Signal Regulated Protein Kinase (ERK) pathways, which promote tumour cell proliferation and survival [12]. HER-3 was reported as one of the most potent oncogenic factors in promoting breast cancer tumourigenesis [13] and is associated with resistance to HER2 inhibitors in HER-2+ breast cancers and anti oestrogen therapy in ER+ breast cancers [14].
The aim of this study was to evaluate HER-3 mRNA expression level as a prognostic marker for breast cancer and to correlate its level with other established prognostic parameters such as stage, grade, lymph node status and molecular subtypes.
Materials and Methods
This study was carried out at Medical Biochemistry and Pathology Departments, Faculty of Medicine. It included 100 female patients. They were classified into two groups, Group I: 40 patients presented with fibroadenoma, and group II: 60 breast patients presented with IDC not otherwise specified and underwent modified radical mastectomy. These cases were received in Pathology Department, in the period between January 2014 and August 2015. Fresh part of the tumour mass was collected in centrifuge tube and kept in -800C. Slices from the tumour mass were then immersed in formalin and was submitted to routine tissue processing ending with paraffin embedded blocks formation. Tumours were graded according to the criteria of Nottingham modification in the Bloom-Richardson system [15]. Tumour staging was performed according to TNM staging system [16].
Quantitative assay of HER-3 mRNA expression using reverse transcriptase polymerase chain reaction technique
Tissue samples were prepared for total RNA isolation using Qiagen RN easy plus Universal Kit from, USA then RNA quality and purity were assured [17] RNA was stored in -80°C till used, then first step-PCR or cDNA synthesis (reverse transcription step) using QuantiTect Reverse Transcription Kit, Qiagen from USA, using Applied Bio systems 2720 thermal cycler (Singapore) for only one cycle as follows: 10 min at 42°C then, 5 min at 95°C to inactivate Reverse Transcriptase and finally for 5 min at 4°C. GAPDH primers were used in RT-PCR reaction as RNA loading control. Second step- PCR or cDNA amplification (real time PCR step): The cDNA was used in SYBR green based quantitative real time PCR for Relative Quantification (RQ) of HER-3 gene expression by SensiFASTTMSYBR Lo-ROX Kit, USA, using the following designed primers (Midland, Texas):1- HER3 Forward primer sequence:5’ GGTGCTGGGCTTGCTTTT3’, Reverse primer 5’ CGTGGCTGGAGTTGGTGTTA 3’and 2-GAPDH Forward primer sequence: 5’ GAAGGTGAAGGTCGGAGTC3’, Reverse primer:5’GAAGATGGTGATGGGATTTC 3’ lastly, data analysis with the Applied Biosystems 7500 software version 2.0.1. The RQ of HER-3 gene expression was performed using comparative ΔΔCt method [18] where the amount of the target (HER-3) mRNA, is normalized to an endogenous reference gene (GAPDH) and relative to a control.
Immunohistochemical Procedure
Several 5 μm thick sections of paraffin embedded blocks were deparaffinized, dehydrated and then placed in Citrate Buffered Saline (CBS) (pH 6.0) and steamed for 20 min. Endogenous peroxidase activity was blocked by incubation with 6% H2O2 in methanol. The primary antibodies used were mouse monoclonal anti human oestrogen receptor alpha, clone 1D5 (ready to use), mouse monoclonal anti human progesterone receptor, clone PgR 636 (ready to use) and HER2/neu, clone 250, diluted 1/50 (Dako, Copenhagen, Denmark). The antibodies were incubated overnight on the slides. Immunoreactivity for Estrogen Receptor (ER) alpha, Progesterone Receptor (PR) and HER2/neu was visualized using Envision+ (DakoCytomation, Glostrup, Denmark) with DAB chromogen as substrate and Mayer’s hematoxylin as counterstain. ER positive breast carcinoma positive for ER, PR and HER2/neu were used as positive control for ER alpha, PR and HER2/neu. For all reagents, negative controls were prepared by substituting the primary antibodies with cross matched isotopes.
Interpretation of Immunostaining
For ER and PR, the assessment of the immunohistochemical staining was evaluated according to the American Society of Clinical Oncology/College of American Pathologist (ASCO/CAP) guidelines [19], in addition to Histo Score (H-score) [20]. For HER-2/neu, immunoreactivity was evaluated according to ASCO/CAP guidelines [21].
Positive HER-2/neu cases were defined as 3+ positivity (> 10% intense and complete membrane staining); however, score 0 or 1+ was considered negative and 2+ cases were excluded.
Immunohistochemical Subtyping
According to the IHC results of ER, PR, HER-2/neu, cases were classified into groups equivalent to molecular subtypes: luminal subtype: ER+ and or PR+, HER-2/neu-; HER-2/neu positive subtype: ER-, PR-, and HER-2/neu +; and lastly, the triple negative (TN) subtype: ER- PR- Her-2 neu [22,23].
Statistical Analysis
Results were collected, tabulated and statistically analyzed by IBM personal computer and statistical package SPSS version 20. Data was expressed into two phases: I – Descriptive: 1) Mean value and Standard Deviation (SD), median, range: for quantitative data; 2) Frequency and percentage for qualitative data. II- Analytic: Mann-Whitney U test for comparison of two independent quantitative variables not normally distributed. K (Kruskal Wallis) test: for comparison of more than two independent quantitative variables not normally distributed. Spearman Correlation Coefficient (r): for comparison between two dependents quantitative not normally distributed variable. The p-value < 0.05 was considered statistically significant.
Results
This study was conducted on a total number of 100 female patients divided into two groups; 40 patients presented fibroadenoma and 60 patients presented with IDC not otherwise specified. The clinicopathological characteristics of malignant cases are presented in [Table/Fig-1]. The malignant cases were classified according to ER, PR and HER2/neu immunohistochemical expression into 35 cases of luminal type, 9 cases showed positive Her-2 neu expression [Table/Fig-2] and 16 cases were triple negative.
Clinicopathological data of studied cases.
| Malignant cases | Control group |
|---|
| Number | Percentage |
|---|
| Age (years) | mean±SD | 47.9±13.4 | 48.6±13.5 |
| median | 48.5 | 46 |
| range | 21-82 | 21-82 |
| Tumour size (cm) | mean±SD | 2.9±1.9 | |
| median | 2 | |
| range | 0.75-8.5 | |
| T stage | 1 | 26 | 43 | |
| 2 | 24 | 40 | |
| 3 | 10 | 17 | |
| 4 | 0 | 0 | |
| N stage | 0 | 16 | 27 | |
| 1 | 22 | 37 | |
| 2 | 14 | 23 | |
| 3 | 8 | 13 | |
| Number of positive lymph node | mean±SD | 5.7±4.5 | |
| median | 5 | |
| range | 1-17 | |
| Type | Luminal | 35 | 58 | |
| Her-2/neu positive | 9 | 15 | |
| triple negative | 16 | 27 | |
| Nearby insitu component | negative | 46 | 77 | |
| positive | 14 | 23 | |
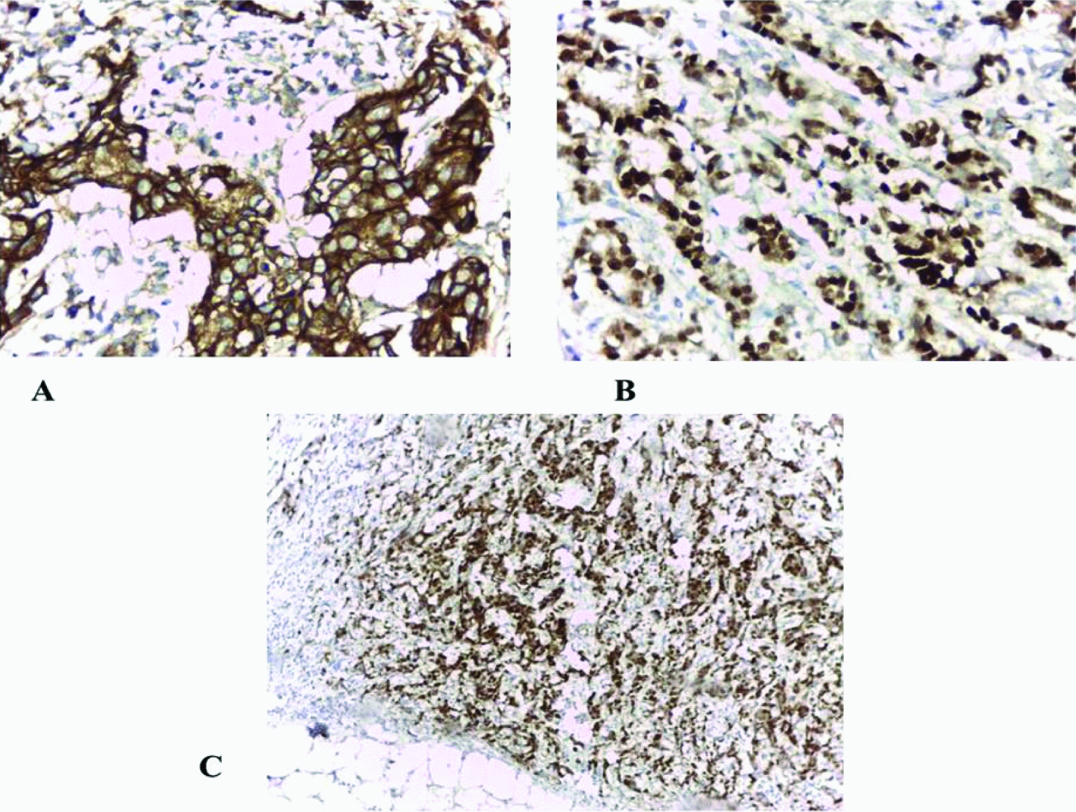
A:Her-2/ neu+strong membranous staining (+++) (immunohistochemical staining x200), B: Strong diffuse nuclear expression for oestrogen receptor (immunohistochemical staining x 400), and C: Strong diffuse nuclear expression for progesterone receptor (immunohistochemical staining x 100).
HER-3 mRNA level in the studied malignant and benign cases.
There was significant difference between benign and malignant breast lesions regarding HER-3 mRNA level, since, its mean and median values were high in malignant cases compared to benign one (p<0.05) [Table/Fig-3,4,5 and 6].
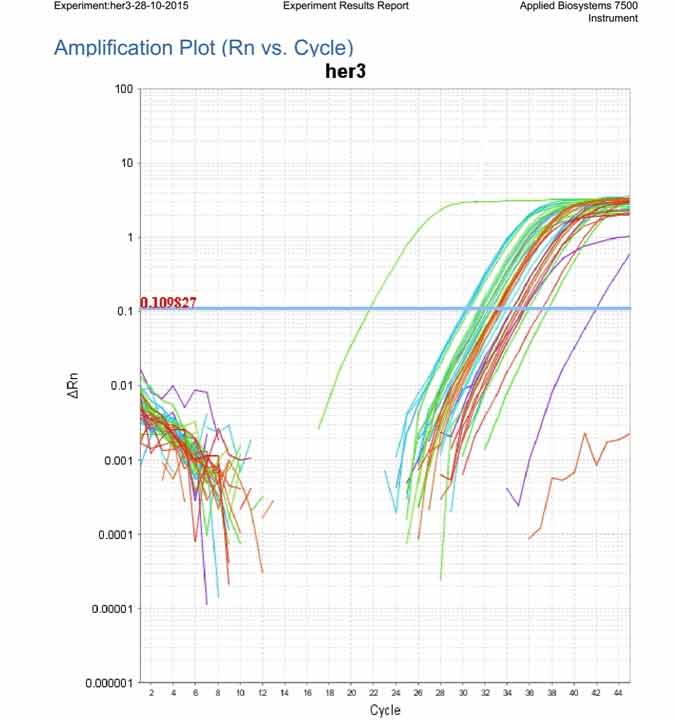
Amplification plot showing the expression of Her3 mRNA in all examined samples.
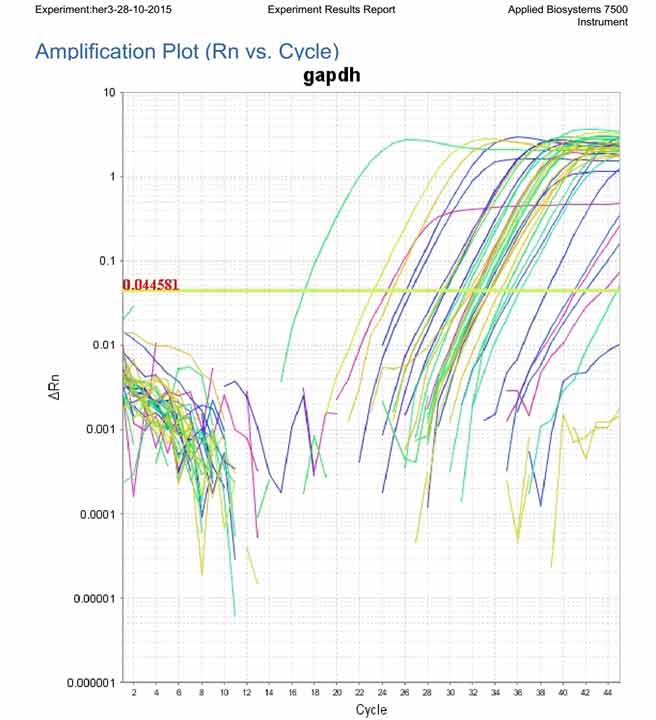
Amplification plot showing the expression of reference gene (GAPDH) in all examined samples.
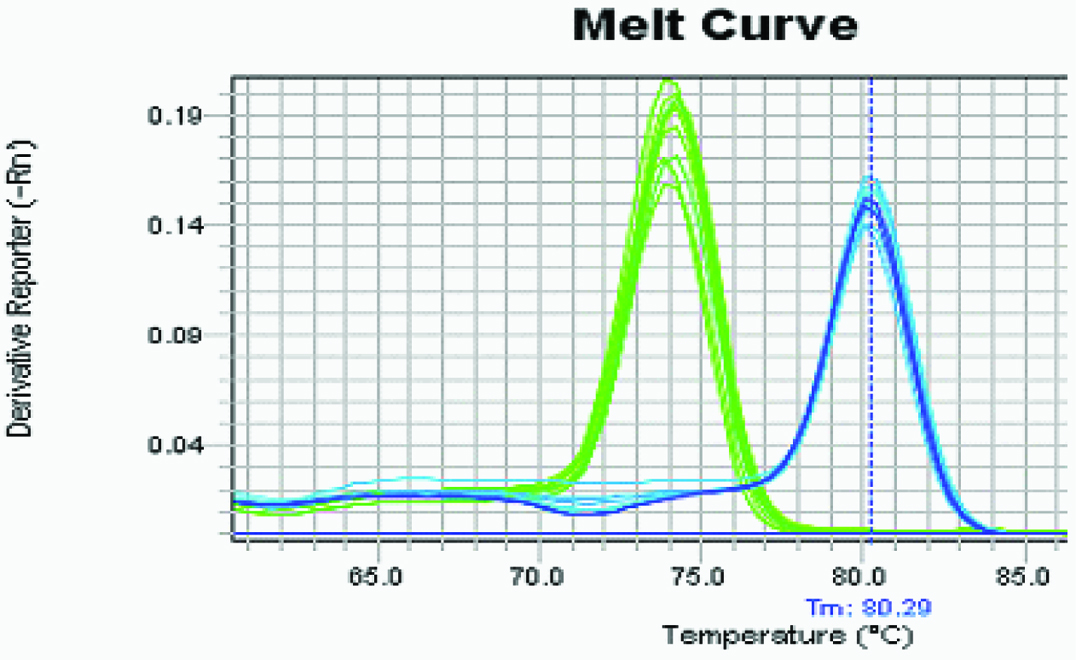
Melting curve analysis identifying specificity of chosen primers.
Comparison between control and malignant groups regarding RQ of HER-3 mRNA expression.
| | Control group | Malignant cases | Testp value |
|---|
| RQ of HER3mRNA | Mean±SD | 0.7±0.6 | 108.5±98.1 | 5.8**<0.05 |
| Median | 0.5 | 80 |
| Range | 0.02-2.8 | 7-411 |
HER-3: human epidermal growth factor receptor 3, **: Mann Whitney test, RQ: relative quantification.
The relationship between HER3 mRNA level and the studied clinicopathological parameters in breast carcinoma
The mean and median values of HER3 mRNA level were significantly associated with breast carcinoma of advanced T stage (p<0.001) and advanced N stage (p<0.001). Furthermore, its level was significantly of highest values in Her-2/neu positive group followed by triple negative cases with the lowest level in luminal group (p<0.05). There was a statistical positive correlation between HER-3 mRNA level and large tumour size (p<0.001) and number of positive lymph nodes (p<0.001). Moreover, breast carcinoma with an adjacent carcinoma in situ showed higher values of HER-3 mRNA level compared to pure cases without in situ component (p<0.05) [Table/Fig-7]. On the other hand no significant association was identified with other parameters such as tumour grade and patient’s age.
Correlation between RQ of HER3 mRNA levels and clinicopathological data of studied malignant cases.
| No. of cases | RQ of HER3 mRNA | Kruskal wallis | p- value |
|---|
| Mean±SD | Median | Range |
|---|
| Age | 60 | | | | -0.03* | 0.8 |
| Tumour size | 60 | | | | 0.92* | <0.001 |
| T stage | early (T1 and T2) | 50 | 77.9±57.4 | 67.5 | 7-228 | 7.5 | <0.001 |
| advanced (T3 and T4) | 10 | 261.4±118.4 | 290.5 | 39-411 |
| N stage | early (N0 & N1) | 38 | 56.5±43.5 | 44 | 7-184 | 7.5 | <0.001 |
| advanced (N2 and N3) | 22 | 198.2±101.9 | 170 | 47-411 |
| Number of positive lymph nodes | 60 | | | | 0.93* | <0.001 |
| Type | Luminal | 35 | 44.3±26.3 | 39 | 7-92 | 107.1 | <0.05 |
| Her2 positive | 9 | 284±78.8 | 267 | 178-411 |
| Triple negative | 16 | 149.9±55.5 | 127 | 89-314 |
| Nearby insitu com-ponent | Negative | 46 | 74.5±72.9 | 57.5 | 7-378 | 6.2 | <0.001 |
| Positive | 14 | 219.9±88.3 | 217 | 89-411 |
*: r (Spearman correlation), HER2: human epidermal growth factor receptor 2, HER3: human epidermal growth factor receptor 3, RQ: relative quantification.
Discussion
The development of breast cancer involves multiple steps of progression, which started with ductal hyper proliferation, followed by carcinoma in situ, invasive carcinoma, and eventually metastatic disease [24]. In human breast cancers both HER-3 mRNA and protein are up regulated. Compared to normal breast tissue, HER3 protein over expression has been reported in 50–70% of human breast cancers and seems to be associated with metastasis, tumour size, and risk of local recurrence [10].
In the present study, there was a significant statistical increase of HER-3 mRNA expression fold change (RQ) in IDC group when compared to benign cases, Kalemi et al., demonstrated similar findings and HER3 was found to exhibit the strongest mRNA expression among other T1GFR family members but instead of benign group, healthy subjects were investigated [25]. Studies utilizing IHC on paraffin embedded tissue have reported HER-3 over expression ranging from 13.8% to 75% of breast cancers [26,27]. Upregulation and reactivation of HER-3 function as escape mechanisms from EGFR inhibition and may play a role in tumour resistance to EGFR targeting tyrosine kinase inhibitors [28].
The current study demonstrated HER-2/neu over expression by IHC in 15% of breast cancer patients (9/60) agreeing with others [29] but it was much lower than other studies [30] who reported 79.5% (39/49). This could be attributed to the increasing aggressiveness of their tumours due to the younger age of their patients, also the staining intensity required to diagnose HER-2/neu over expression and the used antibodies vary among studies.
The current study demonstrated the highest level of HER-3 mRNA expression in HER-2/neu positive cases (aggressive and fast growing) compared to other molecular subtypes such as triple negative and luminal types. The least level of expression was in luminal group (The highest survival rates and fairly low recurrence rates) while higher levels of HER-3 was found in triple negative group (poorer prognosis than luminal one).
Significant coexpression of HER-2 and HER-3 in breast cancer has been inconsistently reported in the literature [31,32]. Formation of dimers between HER-3 and HER-2 seems to be crucial for the HER-2-driven signals in those tumours with HER-2 over expression [4]. Ocana et al., stated that, the expression of HER-3 is associated with worse survival in solid tumours [33]. The influence of HER-3 may be greater in those tumours where HER-2 is commonly over expressed.
HER-3 over expression by immunoprecipitate and immunoblotting protein quantification, RT-PCR, and genomic transfection have been demonstrated in both preclinical invitro and invivo studies to promote breast cancer oncogenesis and tumour growth [30]. The present study demonstrated the association of HER-3 mRNA level with several bad prognostic parameters in breast carcinoma such as advanced T stage, large tumour size and number of positive lymph nodes agreeing with others [27].
In 1992, Lemoine et al., reported the association between HER-3 expression with a worse prognosis [34] and Travis et al., found that tumours with HER-3 expression appeared to develop locally recurrent disease [26]. A Malaysian study by Naidu et al., suggested that it could be involved in the progression of tumours from preinvasive to invasive stage [35]. Furthermore it was associated with established poor prognostic factors. Miller et al., stated that HER-3 is associated with targeted therapeutic resistance to HER-2 inhibitors in HER-2+ breast cancers and anti oestrogen therapy in ER+ breast cancers [14].
Limitation
The limitations of the current study are lacking follow up and survival data together with absence of normal breast tissue to serve as true control.
Conclusion
HER3 gene was upregulated in IDC especially those carrying poor prognostic features. HER-3 mRNA level may identify a subset of patients with a poor prognosis, and who could undergo further evaluation for the efficacy of HER-3 targeted anticancer therapy.
HER-3: human epidermal growth factor receptor 3, **: Mann Whitney test, RQ: relative quantification.
*: r (Spearman correlation), HER2: human epidermal growth factor receptor 2, HER3: human epidermal growth factor receptor 3, RQ: relative quantification.
[1]. Eccles S, Aboagye E, Ali S, Anderson A, Armes J, Berditchevski F, Critical research gaps and translational priorities for the successful prevention and treatment of breast cancerBreast Cancer Res 2013 15:R92 [Google Scholar]
[2]. Soliman A, Hirko K, Hablas A, Seifeldin I, Ramadan M, Banerjee M, Trends in breast cancer incidence rates by age and stage at diagnosis in gharbiah, Egypt, over 10 Years (1999–2008)Journal of Cancer Epidemiology 2013 :1-7. [Google Scholar]
[3]. Olayioye MA, Update on HER-2 as a target for cancer therapy: intracellular signaling pathways of ErbB2/HER-2 and family membersBreast Cancer Res 2001 3:385-89. [Google Scholar]
[4]. Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, Hynes NE, The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumour cell proliferationProcNatlAcadSci U S A 2003 100:8933-38. [Google Scholar]
[5]. Neve RM, Lane HA, Hynes NE, The role of overexpressed HER-2 in transformationAnn Oncol 2001 12(suppl 1):S9-S13. [Google Scholar]
[6]. Kraus M, Issing W, Miki T, Popescu N, Aaronson S, Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: Evidence for over expression in a subset of human mammary tumoursProceedings of the National Academy of Sciences of the United States of America 1989 86:9193-97. [Google Scholar]
[7]. Roskoski R, The ErbB/HER family of protein tyrosine kinases and cancerPharmacol. Res 2014 79:34-74. [Google Scholar]
[8]. Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factorMol Cell Biol 1996 16:5276-87. [Google Scholar]
[9]. Shi F, Telesco S, Radhakrishnan Y, Lemmona M, “ErbB3/HER-3 intracellular domain is competent to bind ATP and catalyze auto phosphorylation,”Proceedings of the National Academy of Sciences of the United States of America 2010 107:7692-97. [Google Scholar]
[10]. Jiang N, Saba N, Chen Z, Advances in targeting HER-3 as an anticancer therapyChemotherapy Research and Practice 2012 2012:1-9. [Google Scholar]
[11]. Hsieh A, Moasser M, Targeting HER proteins in cancer therapy and the role of the non-target HER3British Journal of Cancer 2007 97:453-57. [Google Scholar]
[12]. Amin D, Campbell M, Moasser M, The role of HER3, the unpretentious member of the HER family, in cancer biology and cancer therapeuticsSemin Cell DevBiol 2010 21:944-50. [Google Scholar]
[13]. Rong Ren X, Wang J, Osada T, Mook R, Morse M, Barak L, Perhexiline promotes HER3 ablation through receptor internalization and inhibits tumour growthBreast Cancer Res 2015 17:20 [Google Scholar]
[14]. Miller TW, Pérez-Torres M, Narasanna A, Guix M, Stål O, Pérez-Tenorio G, Loss of phosphatase and tensin homologue deleted on chromosome 10 engages ErbB3 and insulin-like growth factor-I receptor signaling to promote antioestrogen resistance in breast cancerCancer Res 2009 69:4192-201. [Google Scholar]
[15]. Elston C, Ellis I, Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow upHistopathology 1991 19:403-10. [Google Scholar]
[16]. Edge S, Byrd D, Compton C, BreastIn: AJCC Cancer Staging Manual 2010 7th editionNew York, NYSpringer:347-76. [Google Scholar]
[17]. Wang E, Miller L, Ohnmacht G, Liu E, Marincola F, High fidelity mRNA amplification for gene profilingNature Biotechnology 2000 18:457-63. [Google Scholar]
[18]. Dorak M, Real time PCRClinical Chemistry 2004 50:1680-82. [Google Scholar]
[19]. Hammond M, Hayes D, Dowsett M, Allred D, Hagerty K, Badve S, American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of oestrogen and progesterone receptors in breast cancerArch Pathol Lab Med 2010 134:907-22. [Google Scholar]
[20]. Bilalovic N, Berit B, Golouh R, Nesland J, Selak I, Torlakovic E, CD10 protein expression in tumour and stromal cells of malignant melanoma is associated with tumour progressionMod Pathol 2004 17:1251-58. [Google Scholar]
[21]. Wolff A, Hammond M, Schwartz J, Hagerty K, Allred D, Cote R, American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancerJ Clin Oncol 2007 25:118-45. [Google Scholar]
[22]. Sørlie T, Perou C, Tibshirani R, Aas T, Geisler S, Johnsen H, Gene expression patterns of breast carcinomas distinguish tumour subclasses with clinical implicationsProc Natl Acad Sci USA 2001 98:10869-74. [Google Scholar]
[23]. Carey L, Perou C, Livasy C, Dressler L, Cowan D, Conway K, Race, breast cancer subtypes, and survival in the Carolina Breast Cancer StudyJAMA 2006 295:249 [Google Scholar]
[24]. Hirata B, Oda J, Guembarovski R, Ariza C, de Oliveira C, Watanabe M, Molecular markers for breast cancer: Prediction on tumour behaviorDisease Markers 2014 2014:1-12. [Google Scholar]
[25]. Kalemi T, Papazisis K, Lambropoulos A, Voyatzi S, Kotsis A, Kortsaris AH, Expression of the HER family mRNA in breast cancer tissue and association with cell cycle inhibitors p21(waf1) and p27(kip1)Anticancer Res 2007 27:913-20. [Google Scholar]
[26]. Travis A, Pinder S, Robertson J, Bell JA, Wencyk P, Gullick WJ, C-erbB-3 in human breast carcinoma: Expression and relation to prognosis and established prognostic indicatorsBr J Cancer 1996 74:229-33. [Google Scholar]
[27]. Witton C, Reeves J, Going J, Cooke TG, Bartlett JM, Expression of the HER1–4 family of receptor tyrosine kinases in breast cancerJ Pathol 2003 200:290-97. [Google Scholar]
[28]. Mendell J, Freeman D, Feng W, Hettmann T, Schneider M, Blum S, Clinical translation and validation of a predictive biomarker form patritumab, an anti-human epidermal growth factor receptor-3 (HER-3) Monoclonal Antibody, in Patients with Advanced Non-small Cell Lung CancerEBio Medicine 2015 2:264271 [Google Scholar]
[29]. Selim A, El Ayat G, Wells G, C-erbB-2 oncoprotein expression, gene amplification and chromosome 17 aneusomy in breast cancerJ. Pathol 2000 191:138-42. [Google Scholar]
[30]. Yassin D, Kamel A, Taha H, Lotayef M, Hassan Y, Fawzy A, HER family expression in egyptian breast cancer patientsJournal of the Egyptian Nat. Cancer Inst 2003 15:373-80. [Google Scholar]
[31]. Lee Y, Cho S, Seo J, Shin BK, Kim HK, Kim I, Correlated expression of erbB-3 with hormone receptor expression and favorable clinical outcome in invasive ductal carcinomas of the breastAm J Clin Pathol 2007 128:1041-49. [Google Scholar]
[32]. Aubele M, Auer G, Walch A, Munro A, Atkinson MJ, Braselmann H, PTK (protein tyrosine kinase)-6 and HER2 and 4, but not HER1 and 3 predict long-term survival in breast carcinomasBr J Cancer 2007 96:801-07. [Google Scholar]
[33]. Ocana A, Vera-Badillo F, Seruga B, Templeton A, Pandiella A, Amir E, HER-3 overexpression and survival in solid tumours: A meta-analysisJ Natl Cancer Inst 2013 105:266-73. [Google Scholar]
[34]. Lemoine N, Barnes D, Hollywood D, Hughes C, Smith P, Dublin E, Expression of the c-ab-B 3 gene product in breast cancerBr J Cancer 1992 66:1116-21. [Google Scholar]
[35]. Naidu R, Yadav M, Nair S, Kutty M, Expression of c-erbR-3 protein in primary breast carcinomasBr J Cancer 1998 :1385-40. [Google Scholar]