Profile of Tumour Necrosis Factor Alpha -308 G/A Gene Polymorphism in Psoriatic Patients in Karnataka, India
Deepa Rajesh1, Chaitra Chowdappa2, Rajesh Gurumurthy3, A.V. Moideen Kutty4, Sharath Balakrishna5
1 Research Assistant, Department of Cell Biology and Molecular Genetics, Sri Devaraj Urs Academy of Higher Education and Research, Kolar, Karnataka, India.
2 Postgraduate Student, Department of Cell Biology and Molecular Genetics, Sri Devaraj Urs Academy of Higher Education and Research, Kolar, Karnataka, India.
3 Assistant Professor, Department of Dermatology, Sri Devaraj Urs Medical College, Kolar, Karnataka, India.
4 Professor, Department of Cell Biology and Molecular Genetics, Sri Devaraj Urs Academy of Higher Education and Research, Kolar, Karnataka, India.
5 Assistant Professor, Department of Cell Biology and Molecular Genetics, Sri Devaraj Urs Academy of Higher Education and Research, Kolar, Karnataka, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Sharath Balakrishna, Assistant Professor, Department of Cell Biology and Molecular Genetics, Sri Devaraj Urs Academy of Higher Education and Research, Tamaka, Kolar-563103, Karnataka, India.
E-mail: sharath@sduu.ac.in
Introduction
Tumour necrosis factor-alpha (TNFα) gene -308G/A polymorphism (rs1800629) are associated with psoriasis in several populations worldwide. Presently, there is no literature on the status of this polymorphism in the South Indian population.
Aim
To determine the profile of TNFα -308G/A polymorphism among psoriatic patients.
Materials and Methods
This case-control study involved 74 patients with Psoriasis Vulgaris (PsV) and 74 age and gender matched healthy individuals. Patients were recruited from the Department of Dermatology of R.L. Jalappa Hospital and Research Center, Tamaka, Kolar, Karnataka, India, from March 2014 to March 2016. TNFα -308G/A polymorphism was genotyped by Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) method.
Results
The frequency of TNFα -308A allele 7.4% among psoriatic and 8.8% among non-psoriatic individuals. The difference was not statistically significant (p=0.82).
Conclusion
Our results indicate that TNFα gene -308G/A polymorphism is not a significant marker for the risk of developing PsV among South Indian (Karnataka) psoriatic patients.
Genetics, Immunology, Psoriasis vulgaris
Introduction
Psoriasis is a chronic and proliferative inflammatory disorder which clinically presents as raised, erythematous and scaly plaques on the body. Studies have indicated that prevalence of psoriasis in the Indian population is about 0.4-2.8% [1]. Psoriasis is classified on the basis of age of onset as: Type I (age of onset<40 years) and Type II (age of onset >40 years) [2]. Clinically, psoriasis includes several types like vulgaris, guttate, inverse, pustular, erythrodermic and arthritis. Of these types, PsV is seen in nearly 80% of the patients [1].
Psoriasis is a multifactorial disorder involving interaction between immunological, genetic and environmental factors. Immunological component involves interaction between various cell types that produces inflammatory cytokines [3]. Elevated expression of pro-inflammatory cytokines like TNFα, Interleukins (IL) 2,6,8,12 and interferon γ appears to be responsible for initiation, maintenance and recurrence of skin lesions. In addition, reduced expression of anti-inflammatory cytokines like IL1, IL4 and IL10 is considered to represent an insufficient counter-regulatory capacity of the immunological system in psoriasis [3,4].
PsV patients commonly show elevated plasma levels of TNFα [5]. TNFα is a pro-inflammatory cytokine encoded by TNFα gene. It is located on chromosome 6p21.3 in the Major Histocompatibility Complex (MHC) region between MHC class I and II genes. -238G/A and -308G/A are two common Single Nucleotide Polymorphisms (SNPs) present in the promoter region that affects the expression of TNFα gene [6,7]. These two SNPs are associated with several inflammatory disorders that involve elevated expression of TNFα [7]. Studies in several worldwide populations have shown that functional polymorphisms at the -238 and -308 loci are associated with the risk of developing psoriasis [8–10]. The risk of psoriasis increases in the presence of -238A allele while it reduces in the presence of -308A allele. -308A allele is therefore protective in nature [8]. However, a recent study from Pune, India, found that -308A allele increases the risk of developing PsV [11]. The inconsistency of this observation with the worldwide data and lack of information in the South Indian population encouraged us to undertake this study.
Materials and Methods
We adopted case-control design for the study. The study population involved a total of 74 PsV patients and 74 age and gender matched healthy individuals as controls. Inclusion criteria for the selection were patients with erythematous plaques with silvery white micaceous scales. Exclusion criteria were psoriatic patients of subtype other than vulgaris. The control participants did not have a history of psoriasis and other diseases like diabetes, rheumatoid arthritis, ankylosing spondylitis, inflammatory bowel disease and Crohn’s disease. Participants were selected from the Kolar area of Karnataka, India from March 2014 to March 2016. The study was approved by the Institutional Ethics Committee. Participants were enrolled in the study after obtaining informed consent. PsV was diagnosed based on history and clinical examination; PsV was considered as Type I if skin symptoms occurred before 40 years of age, and as Type II if age of onset was more than 40 years. Disease severity was assessed by determining the Psoriatic Area and Severity Index (PASI) [12].
Sample Collection and DNA Isolation: About 2-3 ml of peripheral blood was collected in EDTA vacutainer and stored at 4°C. Genomic DNA was purified from the peripheral blood lymphocytes using salting out method [13]. Purity of the DNA sample was analyzed by UV spectrophotometry (Perkin Elmer model Lambda 35, USA).
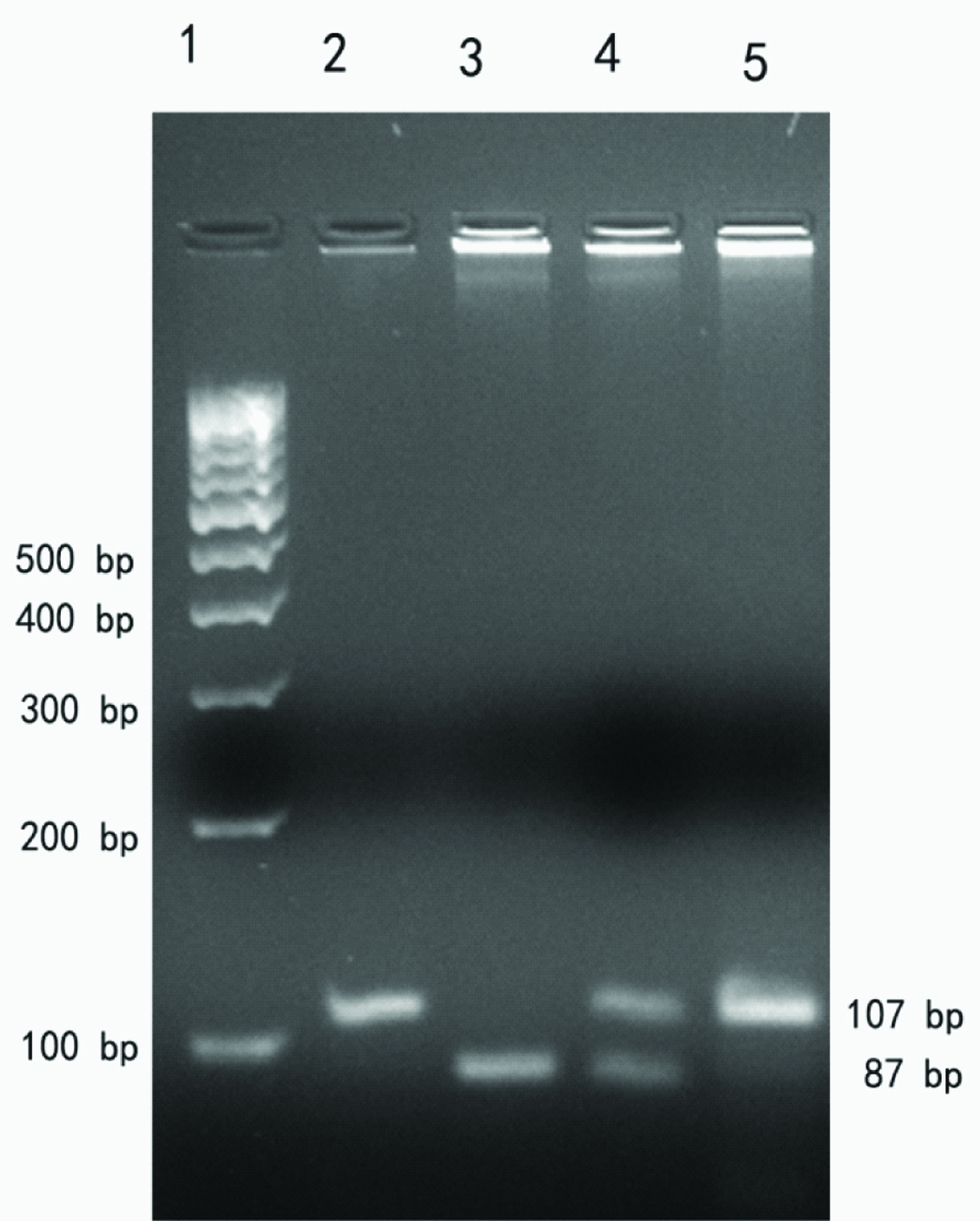
Genotyping TNFα-308G/A: Genomic DNA was amplified by PCR using Bio-Rad C1000 Touch Thermal Cycler. The forward and reverse primer pairs used for the PCR assay were 5’- AGG CAA TAG GTT TTG AGG GCC AT -3’ and 5’- TCC TCC CTG CTC CGA TTC -3’ respectively [14]. Each 25 μl PCR reaction mix comprised of 1x assay buffer, 100 ng genomic DNA, 0.2 mM dNTP, 1 picomole of each primer, 1.5 mM MgCl2 and 1 unit Taq DNA polymerase (Bangalore Genei, India). The following PCR program was used for the amplification reaction: initial denaturation at 95°C for five minutes followed by 35 cycles at 95°C for 30 seconds, 63°C for 30 seconds and 72°C for 30 seconds; final extension involved five minutes at 72°C. The PCR product was analyzed on 3% agarose gel. The 107 bp amplicon was digested with 10 units of NcoI (New England Biolabs, USA) at 37°C and analyzed on 3% agarose gel. The pattern of restriction length polymorphism was analyzed as follows: GG (87+20 bp), AA (107 bp) and GA (107+87+19 bp) [14]. Representative agarose gel image is shown in [Table/Fig-1]. A 10% of the samples were randomly selected for confirmation and the results were 100% concordant. Sanger sequenced samples were used as positive controls.
Representative agarose gel electrophoresis image showing PCR amplicon and RFLP pattern for TNFα -308 G/A SNP. Lane 1, 100bp DNA ladder; Lane 2, PCR amplicon (107 bp); Lane 3, GG genotype (87 bp); Lane 4, GA genotype (107+87 bp); Lane 5, AA genotype (107 bp)
Statistical Analysis
Sample size was calculated using OpenEpi web tool and data from the Pune study [11]. The number of samples required for 95% power was 60 cases and 60 controls. Statistical analysis was done using the Statistical Packages for Social Sciences software (SPSS, version 22, SPSS Inc., Chicago, IL, USA). Contingency table was used to compare the allele and genotype frequencies of the two groups. A p-value was calculated using Fisher’s-exact test. Web program by Rodriguez and co-workers was used to test for conformity to Hardy-Weinberg equilibrium [15]. A p-value <0.05 was considered as statistically significant.
Results
Summary of the clinical profile of the PsV patients included in the study is listed in [Table/Fig-2]. The age of the PsV patients ranged from 22-78 years. Among them 54 (72.9%) were males and 20 (28.7%) were females. The age of the normal healthy individuals included as controls ranged from 20-70 years. Among them 56 (75.7%) were males and 18 (24.3%) were females. The prevalence of Type I and Type II forms was found to be almost similar. Almost 48.6% of the patients showed Type II PsV and 51.3% of the patients showed Type I PsV. In terms of disease severity, 43% of the patients showed mild form and 57% of the patients showed moderate to severe form of PsV.
Demographic and clinical characteristics of cases and controls.
| Parameter | Observation |
|---|
| Gender |
| - Patients (n = 74) | Male: 54 - Female: 20 |
| - Controls (n = 74) | Male: 56 - Female: 18 |
| Age (years) |
| - Patients | 41.0±13.9 |
| - Controls | 42.0±13.9 |
| Clinical type |
| - Type I | 38 |
| - Type II | 36 |
| Severity |
| - Mild | 32 |
| - Moderate -Severe | 42 |
All patients and control subjects were genotyped for the TNFα-308G/A polymorphism. Allele and genotype frequencies in patient and control groups are summarized in [Table/Fig-3]. The distribution of the genotypes in both case and control groups was in agreement with Hardy-Weinberg equilibrium.
Distribution of TNFα -308 G/A alleles and genotypes in the study population.
| Allele/Genotype | Cases(n = 74) | Controls(n = 74) | p-value |
|---|
| G | 137 | 135 | 0.82 |
| A | 11 | 13 |
| GG | 64 | 61 | 0.65 |
| GA | 9 | 13 |
| AA | 1 | 0 |
Allele frequency of -308A among PsV patients was 7.4% and 8.8% in the control group. The distribution of the-308A allele among cases and controls was compared by means of contingency table. A p-value for the distribution profile (p=0.82) was > 0.05 indicating that the two groups did not show any statistically significant difference.
We also compared the distribution of the genotype among patient and control groups. Homozygous major allele (GG) was the predominant genotype seen in both psoriatic and non-psoriatic individuals. The frequency was 86.5% in psoriatic and 82.4% in the non-psoriatic individuals. The minor allele (-308A) was present in very low frequency either in homozygous or heterozygous conditions in both the groups [Table/Fig-3]. The distribution of the genotypes in the two groups was compared by means of contingency table. The two groups did not show any statistically significant difference.
The distribution of the genotypes was stratified on the basis of type of PsV and the data was analyzed for disease association by means of contingency table [Table/Fig-4]. The two subgroups did not show any major difference when they were tested separately. We then stratified the distribution of the genotypes on the basis of severity of PsV and subjected the profile to statistical analysis. Furthermore, no significant association was found either with mild and moderate-severe subgroups.
Distribution of TNFα -308 G/A genotypes in the psoriatic subgroups.
| Genotype | Mild | Moderate-Severe | Type-I | Type-II |
|---|
| GG | 27 | 37 | 34 | 30 |
| GA | 4 | 5 | 6 | 3 |
| AA | 1 | 0 | 1 | 0 |
We saw familial incidence of PsV in four cases. Thus, the frequency of familial incidence was 5.4%. All four patients were male and showed Type I PsV. Of these, one patient showed mild PsV and the remaining three showed severe PsV. Genotype of TNFα-308G/A SNP was GG in two of these patients each and GA in the other two.
Discussion
Elevation of plasma levels of TNFα cytokine in PsV patients is well documented [3]. However, the underlying mechanism that results in the increase is not clear. There are a number of SNPs in the promoter region of TNFα gene which affects the expression and thus, the plasma levels of TNFα [7,16]. The common SNPs that occur in the promoter region of TNFα gene are –238G/A, –308G/A, –857C/T, and –1031T/C. The status of TNFα gene promoter sequence polymorphism in psoriatic patients has been evaluated in several worldwide populations [17,18]. The data from these association studies has been analyzed by at least two meta-analyses [17,18]. Both the meta-analyses found that -238A increased the risk while -308A reduced the risk of developing psoriasis. The two SNPs are assumed to affect the development of psoriasis through separate mechanisms. It is assumed that -238A allele promotes the development of psoriasis by increasing the gene expression of TNFα. In contrast, -308G reduces the gene expression which potentially impairs the clearance of skin infections by pathogens like Candida or Streptococci and thus, predisposes the individual to develop psoriasis [8]. Twin studies indicate that genetic factors influence the development of psoriasis by about 68% [19]. Nearly, 10 genetic loci are indicated to be involved in the genetic predisposition. Of these, PSORS1 and PSORS2 are due to HLACw06 and CARD14 genes respectively [20,21]. These genetic variants show incomplete penetrance. For example, only about 10% of patients who are positive for HLA-Cw6 allele develop psoriasis. This observation indicates the involvement of modifier genes and environmental triggers [22,23]. Genetic variations in TNFα have been an area of intense study since the gene occurs in the HLA class I region [24].
Most of the genetic association studies on TNFα gene are reported from populations that belong to the Caucasian or Mongoloid ethnicity [17]. Literature on the profile of polymorphisms in TNFα gene among Indian psoriatic patients is scanty. The single study by Moorchung NN and co-workers based in Pune (Maharashtra State), India, found that-308A increased the risk of developing psoriasis (p=0.0001; odds ratio=4.38) [11]. The results of this study are in contrast to the meta-analyses of global studies in which -308A is found be protective (odds ratio ~0.5) in role. The conflict in the nature of the association encouraged us to re-examine the frequency in a novel population. The ethnic heterogeneity of the Indian population necessitates testing of association studies in multiple sub-populations. This study is an effort in such a direction. This study did not find any statistically significant difference in the distribution of TNFα-308G/A alleles and genotype between psoriatic and non-psoriatic individuals.
Efforts have been made to explore the role of TNFα-308G/A polymorphism in understanding the pharmacogenetics of anti-TNFα therapeutics. Monoclonal antibodies and recombinant protein bio-similars have been developed to neutralize TNFα with the objective that these molecules can be used as therapeutics in conditions where the cytokine plays a major role. Some of the common examples of this class of molecules are infliximab, adalimumab and etanercept [25]. Studies have shown that TNFα-308G/A polymorphism influences the clinical response to TNFα inhibitors like etanercept, infliximab, and adalimumab [25,26]. In these studies, patients with -308G/G genotype show better response to anti-TNFα therapeutics than those with -308A/A genotype. Similar association has also been noticed on treatment of rheumatoid arthritis patients with infliximab, adalimumab and etanercept [27]. Our data holds significance in the context of pharmacogenetics of anti-TNFα therapeutics. TNFα -308G/G genotype was the predominant genotype seen among psoriatic patients included in this study (86.4%). This observation encourages us to propose that TNFα inhibitors may be a promising therapeutic option for treating psoriasis in our population.
Limitation
Our study is a preliminary explorative study and was conducted in a single center. Further studies involving larger sample size from multiple centers are necessary to confirm our findings.
Conclusion
The results obtained in this study encourage us to conclude that TNFα-308A allele may not be associated with the risk of developing PsV in the South Indian population. The difference of our data with the Pune study and the conclusions of the meta-analyses indicates that the landscape of TNFα-308 G/A polymorphisms among psoriatic patients may be uniquely different. Given the ethnic diversity of India, further studies from multiple study centres rather than larger studies from a single centre would be more useful to understand the association between TNFα-308G/A polymorphisms and predisposition to psoriasis.
[1]. Dogra S, Yadav S, Psoriasis in India: Prevalence and patternInd J Dermatol Venereol Leprol 2010 76(6):595-601. [Google Scholar]
[2]. Krueger G, Ellis CN, Psoriasis – Recent advances in understanding its pathogenesis and treatmentJ Am Acad Dermatol 2005 53:94-100. [Google Scholar]
[3]. Radtke S, Wüller S, Yang XP, Lippok BE, Mütze B, Mais C, Cross-regulation of cytokine signalling: Pro-inflammatory cytokines restrict IL-6 signalling through receptor internalisation and degradationJ Cell Sci 2010 123:947-59. [Google Scholar]
[4]. Ozawa M, Aiba S, Immunopathogenesis of psoriasisCurr Drug Targets Inflamm Allergy 2004 3:137-44. [Google Scholar]
[5]. Kyriakou A, Patsatsi A, Vyzantiadis TA, Sotiriadis D, Serum levels of TNF-α, IL-12/23p40, and IL-17 in plaque psoriasis and their correlation with disease severityJ Immunol Res 2014 2014:467541 [Google Scholar]
[6]. Elahi MM, Asotra K, Matata BM, Mastana SS, Tumor necrosis factor alpha −308 gene locus promoter polymorphism: An analysis of association with health and diseaseBiochimicaet Biophysica Acta 2009 1792(3):163-72. [Google Scholar]
[7]. Qidwai T, Khan F, Tumour necrosis factor gene polymorphism and disease prevalenceScand J Immunol 2011 74(6):522-47. [Google Scholar]
[8]. Kaluza W, Reuss E, Grossmann S, Hug R, Schopf RE, Galle PR, Different transcriptional activity and in vitro TNF-alpha production in psoriasis patients carrying the TNF-alpha 238A promoter polymorphismJ Invest Dermatol 2000 114(6):1180-83. [Google Scholar]
[9]. Arias AI, Giles B, Eiermann TH, Sterry W, Pandey JP, Tumor necrosis factor-alpha gene polymorphism in psoriasisExp Clin Immunogenet 1997 14(2):118-22. [Google Scholar]
[10]. Baran W, Szepietowski JC, Mazur G, Baran E, A-308 promoter polymorphism of tumor necrosis factor alpha gene does not associate with the susceptibility to psoriasis vulgaris. No difference either between psoriasis type I and type II patientsActa Dermatovenerol Alp Pannonica Adriat 2006 15(3):113-18. [Google Scholar]
[11]. Moorchung NN, Vasudevan B, Chatterjee M, Grewal RS, Mani NS, A comprehensive study of tumor necrosis factor-alpha genetic polymorphisms, its expression in skin and relation to histopathological features in psoriasisIndian J Dermatol 2015 60(4):345-50. [Google Scholar]
[12]. Schmitt J, Wozel G, The psoriasis area and severity index is the adequate criterion to define severity in chronic plaque-type psoriasisDermatology 2005 210(3):194-99. [Google Scholar]
[13]. Miller SA, Dykes DD, Polesky HF, A simple salting outprocedure for extracting DNA from human nucleated cellsNucleic Acids Res 1988 16(3):1215 [Google Scholar]
[14]. Guzman-Flores JM, Muñoz-Valle JF, Sánchez-Corona J, Cobian JG, Medina-Carrillo L, Tumor necrosis factor alpha gene promoter −308G/A and −238G/A polymorphisms in Mexican patients with type 2 diabetes mellitusDis Markers 2011 30(1):19-24. [Google Scholar]
[15]. Rodriguez S, Gaunt TR, Day IN, Hardy-Weinberg equilibrium testing of biological ascertainment for mendelian randomization studiesAm J Epidemiol 2009 169(4):505-14. [Google Scholar]
[16]. Hajeer AH, Hutchinson IV, TNF-alpha gene polymorphism: Clinical and biological implicationsMicrosc Res Tech 2000 50(3):216-28. [Google Scholar]
[17]. Li C, Wang G, Gao Y, Liu L, Gao T, TNF-alpha gene promoter -238G>A and -308G>A polymorphisms alter risk of psoriasis vulgaris: A meta-analysisJ Invest Dermatol 2007 127(8):1886-92. [Google Scholar]
[18]. Zhu J, Qu H, Chen X, Wang H, Li J, Single nucleotide polymorphisms in the tumor necrosis factor-alpha gene promoter region alter the risk of psoriasis vulgaris and psoriatic arthritis: A meta-analysisPLoS One 2013 8(5):e64376 [Google Scholar]
[19]. Lonnberg AS, Skov L, Skytthe A, Kyvik KO, Pedersen OB, Thomsen SF, Heritability of psoriasis in a large twin sampleBr J Dermatol 2013 169(2):412-16. [Google Scholar]
[20]. Asumalahti K, Ameen M, Suomela S, Hagforsen E, Michaëlsson G, Evans J, Genetic analysis of PSORS1 distinguishes guttate psoriasis and palmoplantarpustulosisJ Invest Dermatol 2003 120(4):627-32. [Google Scholar]
[21]. Jordan CT, Cao L, Roberson ED, Duan S, Helms CA, Nair RP, Rare and common variants in CARD14, encoding an epidermal regulator of NF-kappaB, in psoriasisAm J Hum Genet 2012 90(5):796-808. [Google Scholar]
[22]. Bhalerao J, Bowcock AM, The genetics of psoriasis: A complex disorder of the skin and immune systemHum Mol Genet 1998 7(10):1537-45. [Google Scholar]
[23]. Bowcock AM, Barker JN, Genetics of psoriasis: The potential impact on new therapiesJ Am Acad Dermatol 2003 49:S51-S56. [Google Scholar]
[24]. Gudjonsson JE, Karason A, Antonsdottir A, Runarsdottir EH, Hauksson VB, Upmanyu R, Psoriasis patients who are homozygous for the HLA-Cw*0602 allele have a 2.5-fold increased risk of developing psoriasis compared with Cw6 heterozygotesBr J Dermatol 2003 148(2):233-35. [Google Scholar]
[25]. Song GG, Seo YH, Kim JH, Choi SJ, Ji JD, Lee YH, Association between TNF-α (-308 A/G, -238 A/G, -857 C/T) polymorphisms and responsiveness to TNFα blockers in spondyloarthropathy, psoriasis and crohn’s disease: A meta-analysisPharmacogenomics 2015 16(12):1427-37. [Google Scholar]
[26]. Murdaca G, Spano F, Contatore M, Guastalla A, Magnani O, Puppo F, Pharmacogenetics of etanercept: Role of TNF-α gene polymorphisms in improving its efficacyExpert Opin Drug Metab Toxicol 2014 10(12):1703-10. [Google Scholar]
[27]. Seitz M, Wirthmuller U, Moller B, Villiger PM, The -308 tumour necrosis factor-alpha gene polymorphism predicts therapeutic response to TNF alpha-blockers in rheumatoid arthritis and spondyloarthritis patientsRheumatology 2007 46(1):93-96. [Google Scholar]