Breast cancer is the fifth most common leading cause of cancer death in women. The burden of breast cancer exceeds all other cancers and the incidence rates of breast cancer are increasing [1]. Breast cancer ranks as number one among all malignant tumors in Egypt accounting for 17.5% and it represents a biologically more aggressive disease than that encountered in the West [2].
Breast cancer is a heterogeneous disease and is classified into three major subtypes: luminal, human epidermal growth factor receptor 2+ (HER2+), and basal like based on comprehensive gene expression profiling [3]. These subtypes differed from each other as regards risk factors for incidence, response to treatment, disease progression, and preferential organ sites of metastases [4].
The proto-oncogene B-cell lymphoma 6 (BCL6) is a master regulator of B-lymphocyte development and facilitates proliferative expansion and blocks differentiation into plasma and memory cells [5]. B cell activation, tolerance of Deoxy Ribonucleic Acid damage, cell cycle arrest, plasma cell differentiation, Nuclear Factor Kappa B (NF-kB) signaling and apoptosis are a broad spectrum of genes modulated by BCL6 [6].
The transcriptional repressor BCL6 contains a N-terminal (BR-C, TTK and bab) (Pox Virus & Zinc finger) BTB/POZ domain and C-terminal zinc finger DNA-binding motifs and represses transcription of a wide range of target proteins and microRNAs. BCL6 is critical for the development, differentiation or function of several cell types [7].
BCL6 binds to specific DNA sequences in the regulatory region of target genes. Once bound, it recruits co-repressor complexes that introduce repressive chromatin marks, thereby repressing transcription [8]. BCL6 also represses genes involved in terminal differentiation as well as its own expression [9].
Both BCL6 and Signal Transducer and Activator of Transcription (STAT) family of transcription factors have overlapping binding sites including STAT3, also known to have an oncogenic role in breast cancer. Triple-negative breast cancer showed complementary roles of BCL6 and STAT3 [10]. STAT5 over STAT3 mediates prolactin-suppression of the BCL6 oncogene in human breast cancer cell lines [11,12].
The aim of this study was to evaluate the expression of BCL6 mRNA in cancer breast and to determine its correlation with the clinico-pathological features including the molecular subtype of breast carcinoma using real time Polymerase Chain Reaction (PCR).
Materials and Methods
This was a prospective case control study, which was carried out at Pathology and Medical Biochemistry Departments, Faculty of medicine, Menoufia University. This study was performed on 100 cases of modified radical mastectomy specimens diagnosed as an invasive duct carcinoma grade II, not otherwise specified (group I) and 50 cases of breast biopsy diagnosed as benign ones (group II). These cases were received in Pathology Department, Faculty of medicine in the period between January 2014 and August 2015. Fresh part of the tumor mass was collected in eppendorf tube and kept in -80° for further RNA extraction and BCL6 mRNA assay. Slices from the tumor mass were then immersed in formalin and was submitted to routine tissue processing ending with paraffin embedded blocks formation. Tumors were graded according to the criteria of Nottingham modification in the Bloom-Richardson system [13]. Tumour staging was performed according to Tumor Node Metastasis (TNM) staging system [14].
Immunohistochemical (IHC) Staining: The method used for immunostaining was streptavidin-biotin–amplified system. From each block, 4μm thick sections were cut on positive charged slides, which were subjected to subsequent steps of deparaffinization, rehydration and antigen retrieval by boiling in citrate buffer saline (pH 6) followed by cooling at room temperature. The primary antibodies were incubated overnight at room temperature and they included Estrogen Receptor (ER) (clone 1D5; Dilution, 1:50) (DakoCytomation), Progesterone Receptor (PR) (clone IA6; Dilution, 1:50) (DakoCytomation) and HER2/neu (clone 250, Dilution, 1:100) (Dako Cytomation). Breast cancer cases positive for ER, PR, and HER2/neu were used as positive control slides. Negative control slides were also included in each run and prepared by the replacement of primary antibodies by buffer solution. Secondary antibody was applied with diaminobenzidine (DAB) as a chromogen substrate and Mayer’s haematoxylin as a counter stain.
Immunostaining Interpretation: ER and PR were considered positive if ≥1% of tumor cell nuclei are immunoreactive [15]. HER2/neu immunoreactivity was evaluated according to the American Society of Clinical Oncology guideline recommendations [16]. Positive HER2/neu cases were defined as 3 positivity (>10% intense and complete staining); however, score 0 or 1 was considered negative.
According to the IHC results of ER, PR and HER2/neu, the cases were classified into;
-Luminal subtype [Table/Fig-1a,b]: positive ER and/or PR and negative HER 2/neu.
Invasive duct carcinoma luminal type showing moderate to strong nuclear immunoreactivity for ER (a) and PR (b). Her 2 neu positive strong membranous staining (+++) (c) (Immunohistochemical staining & x200).
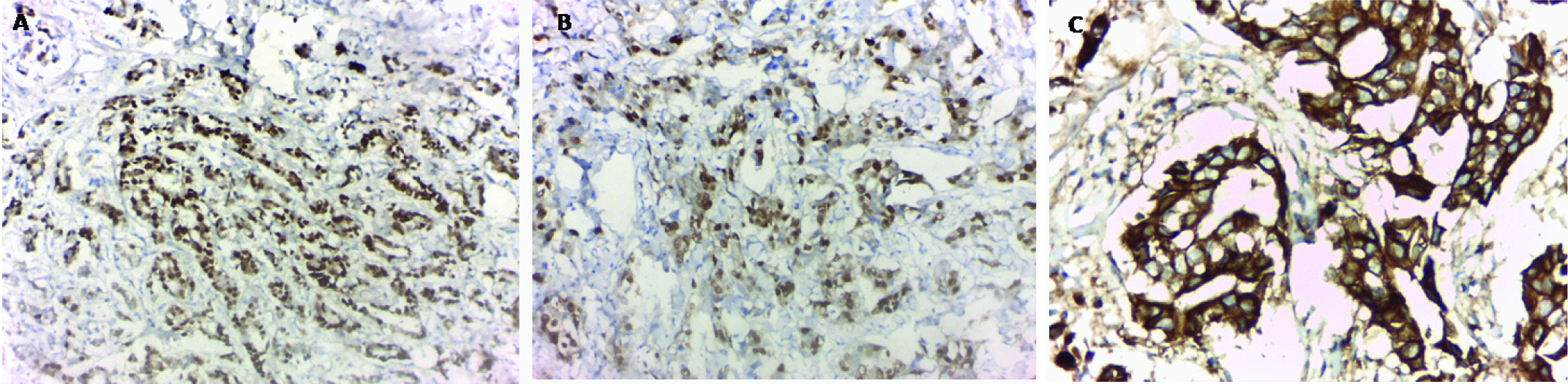
-HER 2/neu positive subtype [Table/Fig-1c]: negative ER, negative PR and positive HER2/neu.
- Triple negative (TN) subtype: negative ER, negative PR and negative HER 2 neu [17].
Assay of BCL6 mRNA
RNA extraction from breast tissue: Total RNA was extracted from specimens using a commercially available kit (QIAamp RNA Blood MiniKit, Qiagen, USA) according to manufacturer’s instructions [18].
The purity of RNA was determined by measuring its absorbance at 260 nm (A260). Absorbance readings should be greater than 0.15 to ensure significance. The ratio between the absorbance value at 260 and 280 nm (A260 /A280) gives an estimate of RNA purity. (A260/A280) ratio greater than 1.6 was accepted. If the purity was lower than 1.5, it required re-extraction [19].
Two-step RT–PCR was done as follows: For reverse transcription step, samples were prepared in a final volume of 20ul containing RT buffer, 5.5mM MgCl2, 500mM each dNTP, 2.5mM random hexamers, 0.4U/mL RNase inhibitor, 1.25U/mL Multi scribe reverse transcriptase (PE Applied Biosystems), and 20ng total RNA. Then the samples were incubated at 25°C for 10 minutes and at 48°C for 30 minutes. Heating to 95°C for 5 minutes inactivated the reverse transcriptase on 2720 thermal cycler Singapore.
For cDNA amplification the sequence of primers of BCL6, forward CCAGCCACAAGACCGTCCAT and CCAGCCACAAGACCGTCCAT as reverse primer according to Genbank NC_000003 were used with SensiFAST™ SYBR® Lo-ROX Kit, Applied Biosystems, 850 Lincoln Centre Drive, Foster City, California 94404, USA, nuclease free water, cDNA in a total reaction volume 25ul and using GAPDH as endogenous control.
Thermal cycling conditions comprised an initial incubation at 50°C for 2 minutes, AmpliTaq gold activation at 95°C for 10 minutes, 40 cycles of denaturation at 95°C for 15 seconds and annealing and extension at 60°C for 1 minute [20].
For relative quantification of the results obtained by Reverse Transcription Polymerase Chain Reaction RT–PCR, The comparative Cycle Threshold (Ct) method was used. Analysis was performed using Applied Biosystems 7500, software version 2.0.1. The point at which the PCR product is first detected above a fixed threshold— termed Ct — was determined for each sample. Each run was completed using melting curve analysis to confirm specificity of the amplification and absence of primer dimmers [Table/Fig-2].
Amplification plot of BCL6 gene expression.
Statistical Analysis
Results were collected, tabulated, statistically analyzed by IBM personal computer and statistical package SPSS version 16 (SPSS Inc. Chicago, Illinois, USA). All data were expressed as mean±SD, number and percent. A p-value of < 0.05 was considered statistically significant.
Results
Clinico-Pathological Data of Breast Carcinoma Cases: The studied cases were all females. The tumor size ranged between 1 and 5 cm in maximal dimension. According to T stage, 35%, 46%, 14% and 5% belonged to T1, T2, T3 and T4 stages, respectively. Twenty four percent of cases lacked lymph node affection while 76% of cases showed lymph node involvement since N1 comprising 34%, N2 31% and N3 11%. According to molecular subtyping, 40% of cases were luminal, 39% were Her2 positive and 21% belonged to triple negative category. Nearby carcinoma in-situ component was identified in 11% of the cases [Table/Fig-3].
Clinico-pathological data.
| Malignant cases | Control group |
|---|
| No. | % |
|---|
| Age (years) | Mean±SD | 48.7±12.9 | 45.2±9.1 |
| Median | 48 | 45 |
| Range | 21-82 | 21-59 |
| Tumor size (cm) | Mean±SD | 3.3±1.3 | |
| Median | 3 |
| Range | 1-5 |
| T stage | 1 | 35 | 35 |
| 2 | 46 | 46 |
| 3 | 14 | 14 |
| 4 | 5 | 5 |
| N stage | 0 | 24 | 24 |
| 1 | 34 | 34 |
| 2 | 31 | 31 |
| 3 | 11 | 11 |
| Number of positive lymph node | Mean±SD | 4.3±3.9 |
| Median | 4 |
| Range | 0-17 |
| Type | Luminal | 40 | 40 |
| Her2 positive | 39 | 39 |
| Triple Negative | 21 | 21 |
| Nearby in situ component | Negative | 89 | 89 |
| Positive | 11 | 11 |
Comparison between BCL6 Level in Benign and Malignant Breast Lesions: There was a significant higher Relative Quantition (RQ) of mRNA level of BCL6 in malignant cases (group I) compared to benign cases (group II) (p<0.001) [Table/Fig-4].
Comparison between control and malignant groups regarding RQ of BCL6 mRNA levels.
| Control group | Malignant cases | Test p-value |
|---|
| RQ of BCL6 mRNA | Mean±SD | 6.9±2.4 | 28±16.2 | U= 3.5 p<0.001 HS |
| Median | 7.1 | 22.5 |
| Range | 2.5-12.4 | 10.9-89.7 |
The Relationship between BCL6 Level and the Studied Clinico-Pathological Parameters in Breast Carcinoma: High BCL6 mRNA levels were significantly associated with cases showing advanced tumor stage (T3 and T4) (p=0.04), triple negative subtype (p<0.001) and with cases associated with nearby carcinoma in situ component (p<0.001) [Table/Fig-5].
Correlation between RQ of BCL6 mRNA levels and clinicopathological data of malignant cases.
| No. of cases | BCL6 gene | Test | p-value |
|---|
| Mean±SD | Median | Range |
|---|
| Age | 100 | | | | r= 0.01 | 0.91 |
| Tumor size | 100 | | | | r= 0.15 | 0.13 |
| T stage | early (T1 & T2) | 81 | 26±13.7 | 22.1 | 12.4-89.7 | u= 536 | 0.04 S |
| advanced (T3 & T4) | 19 | 36.7±22.3 | 26.5 | 10.9-81.9 |
| N stage | early (N0 & N1) | 62 | 27.6±17.3 | 21.1 | 10.9-89.7 | u=1 | 0.23 |
| advanced (N2 & N3) | 38 | 28.6±14.3 | 24.2 | 12.4-76.8 |
| Number of positive lymph nodes | 100 | | | | r= 0.06 | 0.56 |
| Type | Luminal | 40 | 18.2±3.8 | 18.5 | 10.9-30.7 | k= 61.4 | <0.001 HS |
| Her2 positive | 39 | 27.3±10.7 | 24.3 | 16-71.5 |
| Triple negative | 21 | 48±20.9 | 43.7 | 20.5-89.7 |
| Nearby insitu component | negative | 89 | 23.5±8.1 | 21.4 | 10.9-51.3 | U=16 | <0.001 HS |
| positive | 11 | 65±17.8 | 67.5 | 30.7-89.7 |
Discussion
BCL6 has earned a reputation as an oncogene in human cancers by negative regulation of p53 [21,22]. Several lines of evidence suggest that BCL6 may play a role in the pathogenesis of breast cancer. BCL6 has been shown to prevent differentiation of mammary cells [23].
In this study, we observed that BCL6 mRNA was significantly higher in malignant cases compared to control group as noticed by others [24,25]. Walker and his colleges demonstrated that BCL6 is expressed in most breast cancer cells lines and that its genetic locus is amplified in approximately 50% of breast tumor samples and the majority of breast cancer cell lines [26]. Also, BCL6 was upregulated in malignant ovarian epithelial cancer [27].
Through its effects on gene regulation, BCL6 controls cellular processes including cell cycle progression, DNA damage sensing, and protein ubiquitination [28,29]. Moreover, BCL6 is important for breast cancer cell survival [26].
The expression of BCL6 positively associates with the tumor-promoting function of Cyclin D1 Protein (CCND1) and Hypoxia-inducible Factor (HIF1α) in invasive breast cancer [30]. While BCL6 is rarely detected by IHC in normal mammary epithelium, it is commonly expressed in breast cancers, and has been found in 68% of high grade ductal carcinomas [23,30]. In our study, levels of BCL6 mRNA were higher in breast cancer cases presented with advanced T-stage. Pinto and his colleges have similar results but not reach the level of significance [31]. Also, in ovarian carcinoma, the expression levels of BCL6 were upregulated in tumors with advanced International Federation of Gynecology and Obstetrics (FIGO) stage [27].
The correlation between BCL6 upregulation and higher tumour burden in the form of advanced T stage or associated in situ component in our study, suggested that, BCL6 may facilitate tumor progression in breast carcinoma mainly via stimulating tumor growth and tumour invasion, which was supported by the results of in vitro experiments [27,32].
BCL6 was reported to exert a “hit-and-run” mechanism in diffuse Large B Cell Lymphoma (DLBCL), which mean that even transient overexpression of BCL6 can obviously trigger the oncogenicity of DLBCL [33]. Wang and his colleges identified that transient over-expression of BCL6 could significantly upregulate CyclinB1 and CDC25B to accelerate the cell cycle progression, thus promoting the tumour cell proliferation [27].
Triple negative breast cancer, comprise about 20 to 30% of breast tumors and carried poor prognosis and increased mortality due to the lack of specific targeted therapy [34]. In this study, we observed that levels of BCL6 mRNA were significantly higher in triple negative cases compared to luminal and HER2 positive cases. However, in another study, expression levels did not correlate with a specific breast cancer subtype [26].
In spite of BCL6 expression in most breast cancer cell lines, including triple negative breast cancer cell lines [26], triple negative breast cancer cell lines were among the most sensitive to BCL6 inhibition [23]. BCL6 is upregulated by STAT3 [12]. STAT3 activation is restricted largely to triple negative breast cancer cell lines and STAT3 signaling has been shown to be important for the survival of triple negative breast tumors [10]. This may explain the association between BCL6 and triple negative subtype in our study.
Both BCL6 and STAT3 play critical roles, in triple negative breast cancer including promoting survival and epithelial mesenchymal transition(EMT), through modulating largely distinct target genes [34]. Triple negative breast tumors may benefit more strongly from treatment with a BCL6 inhibitor. This may be particularly important as these tumors often become resistant to chemotherapy. Targeting BCL6 with peptidomimetic or small molecule inhibitors kills breast cancer cells alone and in combination with STAT3 or Janus kinase 2 (Jak2) inhibitors [26].
Limitation
The limitation of the present study was the absence of follow up and survival data, which would greatly help in assessing the prognostic value of BCL6
Conclusion
This study concluded that BCL6 over-expression may be used as a biomarker to monitor the invasiveness of tumor and to predict the prognostic risk of patients with cancer breast and the BCL6 inhibitors might be considered as targeted therapy for breast cancer.