Introduction
Theophylline, a methylxanthine drug is indicated to treat asthma and other chronic lung disease (emphysema and chronic bronchitis) [1]. It dilates bronchial smooth muscle (bronchodilator) [2]. Theophyline has narrow therapeutic index, implying that a small dosage change would lead to side effects. The use of theophylline may require therapeutic drug monitoring [2,3]. The side effects of theophylline include: tachycardia, headache, nausea, vomitting and confusion [1].
Several hypothesis were built on the mechanism of action of theophylline. First hypothesis states that theophylline dilates smooth muscle of bronchus and vascular system, reduces airway sensitivity to histamine, methacholine, adenosine and other allergens [1]. The other hypothesis states that theophylline binds to adenosine A2B receptors and blocks adenosine that causes broncho-constriction. Theophylline also inhibits phosphodiesterase (type III and type IV) competitively and activates histone deacetylase to prevent the inflammation [1].
The volume distribution of theophylline is 0.3 to 0.7L/kg with 40% bound to albumin [1]. Most of theophylline (90%) is metabolized in liver [4]. Theophylline is metabolized by CYP1A2 (major) and CYP2E1 (minor) genes [5]. CYP1A2 catalyzes the demethylation and hydroxylation of theophylline, while CYP2E1 catalyzes its hydroxylation [6–8].
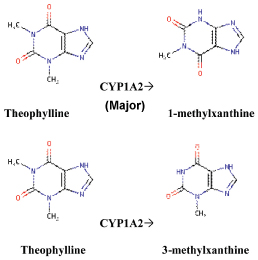
The metabolism of theophylline by CYP1A2 results in metabolite 1 methylxanthin (3-N-demethylation and 3-methyl xantin (1-N-demethylation reaction) [Table/Fig-1].
The metabolism of theophylline by CYP1A2 enzyme [1].
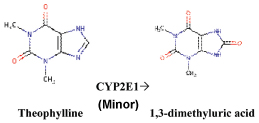
Theophylline is also metabolized (minor) by CYP2E1 enzyme into 1,3-dimethyluric acid (8-hydroxylation reaction) [Table/Fig-2].
The metabolism of theophylline by CYP2E1 [1].
The t1/2 (half life) of theophylline is 8 hours and its clearance is about 1.7mL/kg/min in children (1-4 years); 0.65mL/kg/min in adults (16-60 years) and 0.41mL/kg/min in elderly (>60 years) [1].
Polymorphism
The treatment response is influenced by many factors. Genetic variation is one of the factors that may affect treatment response. Polymorphism is defined as a genetic variation in DNA sequences occurring with frequency of at least 1% in a population. In relation with drug response, polymorphism may occur in some places, such as: polymorphism of the gene encoding the enzymes that metabolize drugs, drug transporter genes and polymorphism in drug target genes [9].
The most common polymorphism is genetic polymorphism of drug metabolizing enzyme. Most polymorphism in phase 1 metabolism occurs by polymorphism at CYP, while in phase II metabolism, the polymorphism occurs by the enzyme N-acetyltransferase, thiopurine S-methyltransferase, and glutathione transferase [9].
Cytochrome P450 (CYP)
Cytochrome P450 (CYP) is an enzyme involved in the metabolism of most drugs. CYP plays an important role in the occurrence of variations in drug response between individuals. There are 57 human CYPs identified, which are classified into 18 families and 43 sub families [10]. Many drugs, xenobiotics and endogenous molecules were metabolized by CYP families 1, 2 or 3 [11]. One CYP can metabolize several drugs and instead, one drug is often metabolized by several CYPs. Several drugs can induce or inhibit activity of CYP [12].
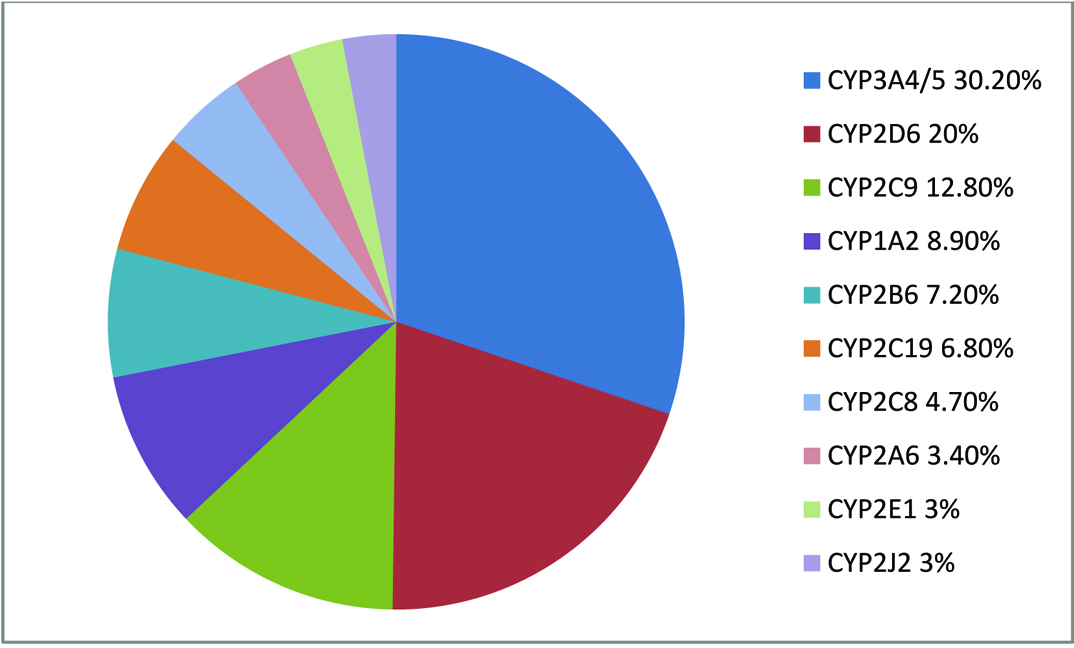
Most drugs are metabolized by CYP3A4/5. The Percentage of drugs metabolized by CYP can be seen in [Table/Fig-3] [13].
The frequency of drugs which were metabolized by CYP [13].
Polymorphism of CYP1A2 gene
The human CYP1A2 gene is located on chromosome 15q24.1. This gene contains 7 exons and 6 introns with molecular weight 58,294 Da [14]. The structural characteristics of this enzyme are aromatic, polyaromatic, heterocyclic amide and amine. The substrates of this enzyme are : caffeine, clomipramine, clozapine, phenacetin, propanolol, theophylline, tizanidine [13,14]. Several drugs are known as inhibitor of this enzyme: cimetidine, ciprofloxacin, disulfiram, mexiletine, tolfenamic acid and oral contraceptives. Some drugs act as potent inducer of CYP1A2. They are: antipyrine, carbamazepine, nelfinavir, omeprazole, phenobarbital, phenytoin, primaquine, rifampicin, ritonavir, sulfinpyrazone [13].
The research conducted by Uslu et al., (2010) on Turkish people found that theophylline is metabolized by CYP1A2. There are several polymorphism genes of CYP1A2, among others: CYP1A2*1C, CYP1A2*1D, CYP1A2*1E and CYP1A2*1F polymorphisms [15]. There are several variant types of CYP1A2 in Japanese population: CYP1A2*1F allele (-163C>A (with a 0.628 frequency, while CYP1A2*15 allele (125C>G), CYP1A2*16 allele (1130G>A) and CYP1A2 *8 allele (1367G>A) with frequencies of 0.002, 0.002 and 0.004, respectively [16]. Wang et al., 2013 stated that there were four polymorphisms of CYP1A2 among Chinese population: CYP1A2*1C (G-3860A), G-3113A, CYP1A2*1F (C-163A) and CYP1A2*1B (C-5347T). The CYP1A2*1F polymorphism increases CYP1A2 activity in subjects with -3860G/-3113G/5347C homozygote (0.66 ± 0.24 versus 0.46 ± 0.05, p = 0.034). The activity of CYP1A2 on people with the -3113 AA genotype was lower than people with the -3113 AG genotype (0.35 ± 0.04 versus 0.48 ± 0.07, p = 0.016) or the -3113 GG genotype (0.35 ± 0.04 versus 0.58 ± 0.22, p = 0.037) [17]. Research by Yoon et al., 2006 stated that the 1,3-dimethyluric acid/theophylline plasma was affected by CYP1A2 -2964G>A gene polymorphism [18].
Polymorphism CYP2E1 Gene
CYP2E1 gene is located on chromosome 10q26.3. This gene encodes enzyme involved in metabolism of theophylline. Almost 7% of total CYPs are CYP2E1 [19]. This enzyme is characterized by being small, hydrophilic and planar. The substrates of this enzyme are: aniline, arachidonic acid, halothane, salicylic acid. Many drugs may inhibit CYP2E1 enzymes: clomethiazole, disulfiram, orphenadrine. However, acetone, ethanol, isoniazid and pyrazole induce this enzyme [13].
There were 11 polymorphisms of CYP2E1 gene in Korean people: Ins(96), -1566 T>A, -1515 T>G, -1414 C>T, -1295 G>C, -1055 C>T, -1027 T>C, -930 A>G, -807 T>C, -352 A>G, and -333 T>A [18]. The frequency of CYP2E1 *1A/*1A in non-white and white among Brazilian population were 96.0% and 87.4%, while CYP2E1 *1A/*5B 2.7% and 11.3%. The frequency of CYP2E1 *5B/*5B, CYP2E1 *1A/*6 and CYP2E1 *6/*6 were 1.3; 1.3; 12.0; 15.2; 1.3 and 0.7 % respectively [20].
The Polymorphism of CYP1A2 and CYP2E1 Genes and Theophylline Response
Previous studies identified that there were varying expressions of CYP1A2 and CYP2E1 genes, at inter-individual level impacting metabolism of several drugs including theophylline [21–25].
The impact of polymorphism of CYP1A2 and CYP2E1 can be seen in [Table/Fig-4] [5,18,26,27].
The impact of polymorphism of CYP1A2 & CYP2E1 on theophylline effect.
| Polymorphism | Type of Polymorphism | Impact on theophylline response | Ref. |
|---|
| CYP1A2 | 3860AG>A | Higher clearance of theophylline on GA&AA allele than GG allele | [5] |
| 3113G>A (CYP1A2*1F) | Decrease of CYP1A2 activity→decrease of theophylline metabolism | [26] |
| 163C>A (CYP1A2*1F) | Increase of CYP1A2 activity →Increase of theophylline metabolism | [27] |
| CYP2E1 | -1055C>T | Decrease CYP2E1 enzyme activity and decrease 1.3-DMU/theophylline ratio | [18] |
| -1027C>T |
| -807T>C |
| -1566T>A |
| -1295G>C |
From [Table/Fig-4] it can be seen that the polymorphism of CYP1A2 -3860G>A point mutation with an A mutant allele genotype (GA+AA) on Korean people have higher theophylline clearance than GG type [5]. Obase et al., (2003), found that there was a significant increase in the theophylline clearance in people with A mutant alleles in contrast with wild type (29.11 ± 0.91mL/kg/h vs. 26.12 ± 0.80 mL/kg/h, p = 0.014). The metabolism of theophylline is lower among asthma people with A allele at -3860G>A [28], while research by Yim showed that no association of theophylline clearance in six SNPs in CYP1A2 (p > 0.05) including -3598G>T [5]. The CYP1A2 polymorphisms (T allele at -2467 del T and the C allele at -163 C >A) increase the risk of chronic obstructive pulmonary disease [15]. Chen et al., (2005) conducted a research on Chinese people and found that the G-3113A polymorphism (CYP1A2*1F) is associated with the decrease of CYP1A2 activity [26], while the *1F allele (CYP1A2*1F (-163C>A) allele) increase of enzyme activity resulting in increased metabolism of theophylline [27].
A significantly higher activity was observed in CYP1A2*1C, CYP1A2*1K, CYP1A2*3, CYP1A2*4, CYP1A2*6, CYP1A2*7, CYP1A2*8, CYP1A2*11, CYP1A2*15, and CYP1A2*16 variants [29–33].
Yoon et al., (2006) found that five polymorphisms of CYP2E1 influenced 1,3-dimethyluric acid (DMU)/theophylline ratio on Korean people. The five SNPs were −1055 C>T; −1027 T>C; −807 T>C; −1566 T>A and −1295 G>C. They reduced CYP2E1 enzyme activity. The 1,3-DMU/ theophylline ratio of each types were as follows: genotypes of CC on 1055 C>T is 0.0533±0.0343 (CT= 0.0368±0.0171); TT on −1027 T>C is 0.0533±0.0343 (TC=0.0368±0.0171); TT on −807 T>C is 0.0533±0.0343 (TC=0.0368±0.0171); TT on −1566 T>A is 0.0533±0.0343(TA=0.0368±0.0171); GG on −1295 G>C is 0.0533±0.0343 (GC=0.0368±0.0171) respectively [18].
Conclusion
Polymorphism of CYP1A2*1F allele (CYP1A2*1F (-163C>A) allele) increases CYP1A2 enzyme activity. The five SNPs of CYP2E1 namely −1055 C>T; −1027 T>C; −807 T>C; −1566 T>A and −1295 G>C reduce CYP2E1 enzyme activity and 1,3-DMU/ theophylline ratio.
[1]. Drug bank. Theophylline. http://www.drugbank.ca/drugs/DB00277 [Google Scholar]
[2]. National Asthma Education and Prevention Program Expert Panel. Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma. U.S. Department of Health and Human services. National Institute of Health, 2007, Pp:240-250 [Google Scholar]
[3]. Mitenko PA, Ogilvie RI, Rational intravenous doses of theophyllineN Engl J Med 1973 289:600-03. [Google Scholar]
[4]. Haley TJ, Metabolism and pharmacokinetics of theophylline in human neonates, children, and adultsDrug Metab Rev 1983 14(2):295-335. [Google Scholar]
[5]. Yim EY, Kang HR, Jung JW, Shhn SW, Cho SH, CYP1A2 polymorphism and theophylline clearance in korean non-smoking asthmaticsAsia Pac Allergy 2013 3(4):231-40. [Google Scholar]
[6]. Robson RA, Miners JO, Matthews AP, Stupans I, Meller D, McManus ME, Characterisation of theophylline metabolism by human liver microsomes. Inhibition and immunochemical studiesBiochem Pharmacol 1988 37:1651-59. [Google Scholar]
[7]. Grygiel JJ, Wing LM, Farkas J, Birkett DJ, Effects of allopurinol on theophylline metabolism and clearanceClin Pharmacol Ther 1979 26:660-67. [Google Scholar]
[8]. Rasmussen BB, Maenpaa J, Pelkonen O, Loft S, Poulsen HE, Lykkesfeldt J, Selective serotonin reuptake inhibitors and theophylline metabolism in human liver microsomes: potent inhibition by fluvoxamineBr J ClinPharmacol 1995 39:151-59. [Google Scholar]
[9]. Cavallari LH, Lam YWF, DiPiro JT, Talbert LI, Yee GC, Matzke GR, Pharmacotherapy: A Pathophysiologic approach 2005 Mc Grawhill:75-90. [Google Scholar]
[10]. Ingelman-Sundberg M, The human genome project and novel aspects of cytochrome P450 research. ToxicolAppl Pharmacol 2005 207:52-6. [Google Scholar]
[11]. McGraw J, Waller D, Cytochrome P450 variations in different ethnic populationsExpert Opin Drug Metab Toxicol 2012 8:371-82. [Google Scholar]
[12]. Flockhart DA, Oesterheld JR, Cytochrome P450-mediated drug interactionsChild Adolesc Psychiatr Clin N Am 2000 9:43-76. [Google Scholar]
[13]. Zanger UM, Schwab M, Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variationPharmacol Ther 2013 138:103-41. [Google Scholar]
[14]. Zhou SF, Wang B, Yang LP, Liu JP, Structure, function, regulation and polymorphism and the clinical significance of human cytochrome P450 1A2Drug Metab Rev 2010 42(2):268-354. [Google Scholar]
[15]. Uslu A, Ogus C, Ozdemir T, Bilgen T, Tosun O, Kesr I, The effect of CYP1A2 gene polymorphisms on theophylline metabolism and chronic obstructive pulmonary disease in Turkish patientsBMB Rep 2010 43(8):530-34. [Google Scholar]
[16]. Soyama A, Saito Y, Hanioka N, Maekawa K, Komamura K, Kamakura S, Single nucleotide polymorphisms and haplotypes of CYP1A2 in a Japanese population. Drug MetabPharmacokinetic 2005 20(1):24-33. [Google Scholar]
[17]. Wang L, Zh H, Deng X, Wang Y, Zhang Z, Cheng Z-N, Association between Common CYP1A2 Polymorphisms and Theophylline Metabolism in Non-smoking Healthy VolunteersBasic Clin Pharmacol Toxicol 2013 112:257-63. [Google Scholar]
[18]. Yoon Y, Park HD, Park KU, Kim JQ, Chang YS, Song J, Associations between CYP2E1 promoter polymorphisms and plasma 1,3-dimethyluric acid/theophylline ratiosEur J Clin Pharmacol 2006 62(8):627-31. [Google Scholar]
[19]. Makowski GS, Advances in in clinical chemistry vol 71 2015 First editionLondonElseiver:92-94. [Google Scholar]
[20]. Rossini S, Soares Lima DCM, Rapozo M, Faria RM, Pinto R, CYP2A6 and CYP2E1 polymorphisms in a brazilian population living in rio de janeiroBraz J Med Biol Res 2006 39:195-201. [Google Scholar]
[21]. Hammons GJ, Guengerich FP, Weis CC, Beland FA, Kadlubar FF, Metabolic oxidation of carcinogenic arylamines by rat, dog, and human hepatic microsomes and by purified flavin-containing and cytochrome P-450 monooxygenasesCancer Res 1985 45:3578-85. [Google Scholar]
[22]. Minchin RF, McManus ME, Boobis AR, Davies DS, Thorgeirsson SS, Polymorphic metabolism of the carcinogen 2-acetylaminofluorene in human liver microsomesCarcinogenesis 1985 6:1721-24. [Google Scholar]
[23]. Watanabe J, Hayashi S, Kawajiri K, Different regulation and expression of the human CYP2E1 gene due to the RsaI polymorphism in the 5’-flanking regionJ Biochem 1994 116:321-26. [Google Scholar]
[24]. Ikeya K, Jaiswal AK, Owens RA, Jones JE, Nebert DW, Kimura S, Human CYP1A2: sequence, gene structure, comparison with the mouse and rat orthologous gene, and differences in liver 1A2 mRNA expressionMol Endocrinol 1989 3:1399-408. [Google Scholar]
[25]. Butler MA, Guengerich FP, Kadlubar FF, Metabolic oxidation of the carcinogens 4-aminobiphenyl and 4,4’-methylene-bis(2- chloroaniline) by human hepatic microsomes and by purified rat hepatic cytochrome P-450 monooxygenasesCancer Res 1989 49:25-31. [Google Scholar]
[26]. Chen X, Wang L, Zhi L, Zhou G, Wang H, Zhang X, The G-113A polymorphism in CYP1A2 affects the caffeine metabolic ratio in a Chinese populationClin Pharmacol Ther 2005 78(3):249-59. [Google Scholar]
[27]. Zhou SF, Yang LP, Zhou ZW, Liu YH, Chan E, Insights into the substrate specificity, inhibitors, regulation, and polymorphisms and the clinical impact of human cytochrome P450 1A2AAPS J 2009 11(3):481-94. [Google Scholar]
[28]. Obase Y, Shimoda T, Kawano T, Saeki S, Tomari SY, Mitsuta-Izaki K, Polymorphisms in the CYP1A2 gene and theophylline metabolism in patients with asthmaClin Pharmacol Ther 2003 73:468-74. [Google Scholar]
[29]. Nakajima M, Yokoi T, Mizutani M, Kinoshita M, Funayama M, Kamataki T, Genetic polymorphism in the 5’-flanking region of human CYP1A2 gene: effect on the CYP1A2 inducibility in humansJ Biochem 1999 125:803-08. [Google Scholar]
[30]. Sachse C, Brockmöller J, Bauer S, Roots I, Functional significance of a C→A polymorphism in intron 1 of the cytochrome P450 CYP1A2 gene tested with caffeineBr J Clin Pharmacol 1999 47:445-49. [Google Scholar]
[31]. Aklillu E, Carrillo JA, Makonnen E, Hellman K, Pitarque M, Bertilsson L, Genetic polymorphism of CYP1A2 in Ethiopians affecting induction and expression: characterization of novel haplotypes with single-nucleotide polymorphisms in intron 1Mol Pharmacol 2003 64:659-69. [Google Scholar]
[32]. Allorge D, Chevalier D, Lo-Guidice JM, Cauffiez C, Suard F, Baumann P, Identification of a novel splice-site mutation in the CYP1A2 geneBr J Clin Pharmacol 2003 56:341-44. [Google Scholar]
[33]. Murayama N, Soyama A, Saito Y, Nakajima Y, Komamura K, Ueno K, Six novel nonsynonymous CYP1A2 gene polymorphisms: catalytic activities of the naturally occurring variant enzymesJ Pharmacol Exp Ther 2004 308:300-06. [Google Scholar]