Epigenetics and Periodontitis: A Contemporary Review
Geetha Ari1, Sandhya Cherukuri2, Ambalavanan Namasivayam3
1 Reader, Department of Periodontology, Meenakshi Ammal Dental College and Hospital, Chennai, Tamil Nadu, India.
2 Postgraduate Student, Meenakshi Ammal Dental College and Hospital, Chennai, Tamil Nadu, India.
3 Professor and Head, Department of Periodontology, Meenakshi Ammal Dental College and Hospital, Chennai, Tamil Nadu, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Geetha Ari, Reader, Department of Periodontology, Meenakshi Ammal Dental College and Hospital, Alapakkam Main Road, Maduravoyal, Chennai-600 095, Tamil Nadu, India.
E-mail: rashmetachn@yahoo.com
Periodontitis is characterized by infection and inflammation of the tooth supporting structures. Few signs of susceptibility and recurrence after treatment are seen due to the outgrowth of various pathogenic microorganisms. Many studies have been done to understand the genetic basis of periodontal disease. An increased risk for periodontitis has been shown with the variations in genes related to the inflammatory response. Interestingly, some of the genes regulated by epigenetic modifications are modified in response to environmental stimuli. Conditions such as cancer, autoimmune or inflammatory diseases have been dispensed by epigenetic mechanisms. The understanding of these molecular mechanisms and the early detection of susceptibility may guide in future periodontal disease treatment and prevention.
DNA methylation, Future implications, Histone modification
Introduction
Periodontitis is a complex disease characterized by inflammation and destruction of tooth supporting structures. Although the definitive mechanism remains unclear, a contributing factor for inflammation and infection is the outgrowth of multiple opportunistic microorganisms in the oral cavity including Porphyromonas gingivalis, Tannerella forsythia and Aggregatibacter actinomycetemcomitans that triggers the host immune response [1]. But the aetiology involves various other extrinsic and intrinsic factors such as genetics and not merely these specific microorganisms that cause the disease [2]. A specific gene possesses different epigenetic patterns depending on the cell type which results as a local and systemic expression of the gene. This review focuses the importance of genetic factors in the progression of periodontal disease and highlight how epigenetic alterations play a role as mediators between genetic and environmental factors. Further research in future will provide better guidelines for the development of novel biomarkers that will aid in the prevention of periodontal disease.
Epigenetics
In Cold Spring Harbor Meeting 2008, Epigenetics was defined as a “stably heritable phenotype resulting from changes in a chromo-some without alterations in the DNA sequence” [3]. In Greek, prefix ‘epi-’ in epigenetic means ‘on the top of’ or ‘in addition to’ genetics. Epigenetic modifications include chemical alterations of DNA and associated proteins, leading to remodelling of the chromatin and activation or inactivation of a gene. These changes can lead to the development and maintenance of cancer and autoimmune or inflammatory diseases, including periodontitis.
The mechanisms underlying epigenetic alterations involve DNA methylation, histone modification and gene regulation by non-coding RNAs [4–6]. These are potentially reversible and transient. It can be induced or altered by environmental factors that modulate the gene expression and affect various gene functions [Table/Fig-1]. Therefore, it produces a link between the inherited genome and the environment [4,7].
Epigenetic alterations affecting various gene functions.
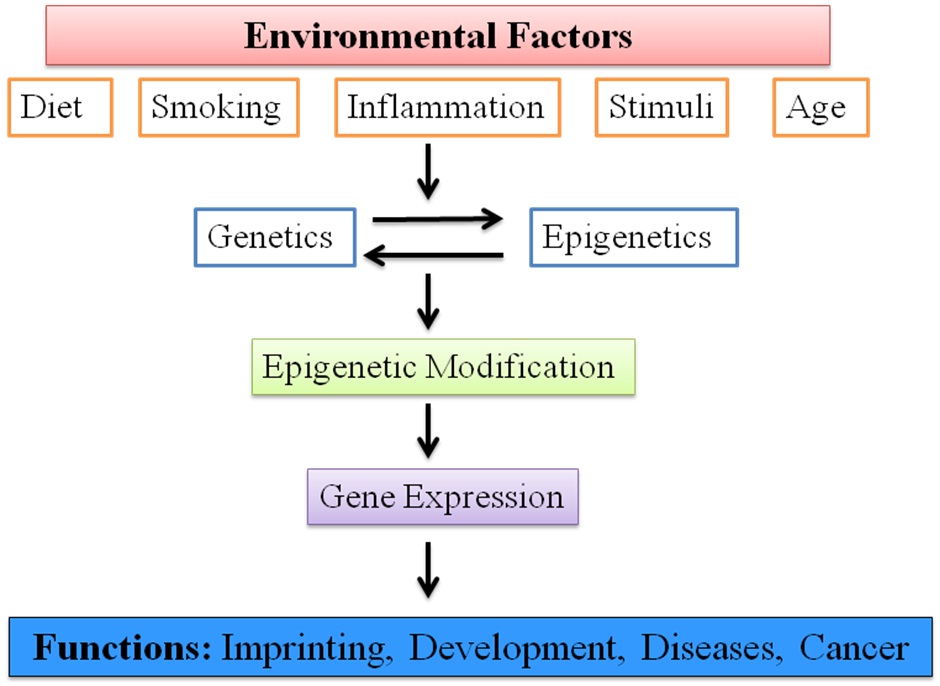
In dentistry though the use of these epigenetic mechanisms is still at the early stage, they play an important role during development and pathological diseases of the oral cavity.
Epigenetics in Periodontitis
Epigenetic mechanisms result in heritable modifications in the expression of genes that are independent from DNA coding variability. The post-translational modification of histone proteins in chromatin and the methylation of DNA are the two primary epigenetic mechanisms in periodontitis. These are regulated by distinct, but coupled, pathways [7].
DNA Methylation: Within the nucleus, chromosomal DNA is tightly associated with proteins, and these interactions form the ordered structure known as chromatin. The methylation takes place in Cytosine-phosphate-Guanine (CpG) dinucleotides of the DNA chain with the covalent transfer of a methyl group from S-Adenosyl Methionine (SAM) to cytosine [Table/Fig-2a] [8]. CpG sequences tend to be germline-specific or associated with imprinted genes, and the methylated DNA sequence in CpG sites cause the more condensed DNA structure. This process leads to transcriptional repression and gene silencing [Table/Fig-2b] [9].
DNA methylation and transcriptional inactivation of gene.
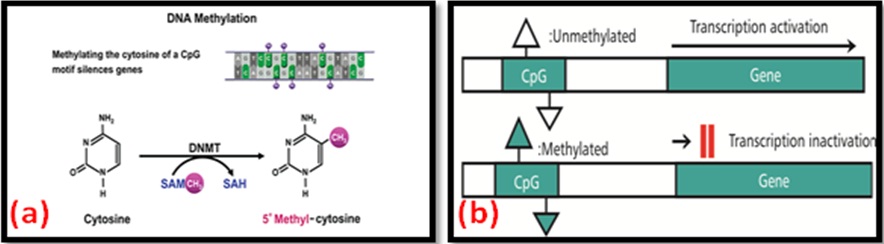
In periodontitis, epigenetic modifications during inflammation occur locally at the biofilm-gingival interface around the teeth. It has been shown that the epigenome differs between inflamed periodontal sites and non-inflamed sites in the same individual [10] and also different methylation patterns associated with pathways regulating cell differentiation, apoptosis, Lipopolysaccharide (LPS) mediated signalling, oncogenesis and cell adhesion were found in inflamed tissue of periodontitis patients compared with tissue from healthy individuals.
[Table/Fig-3] shows few studies that have investigated the process of DNA methylation of inflammatory cytokines in Chronic Periodontitis (CP) and Aggressive Periodontitis (AgP) [11–14].
Various studies of DNA methylation process.
| STUDY | MARKER | RESULTS |
|---|
| Andia et al., 2010 | Interleukin (IL-8) | The IL-8 promoter was hypomethylated in oral epithelial cells of generalized AgP patients when compared with healthy controls. |
| Zhang et al., 2010 | Interferon (IFN–γ) | CpG sites reported hypomethylation and increased expression of INF-γ in periodontitis biopsies compared with healthy tissue biopsies. |
| Zhang et al., 2013 | Tumor Necrosis Factor (TNF-α) | It was found to be hypermethylated in CP. |
| Stefani et al., 2013 | IL-6 | IL-6 promoter found to be partially methylated in both healthy and periodontitis patients, even though increased expression was seen in periodontitis patients. |
AgP: Aggressive Periodontitis; CP: Chronic Periodontitis
Further to methylation level of cytokines, a number of other genes related to inflammation have been assayed in periodontitis. A hypermethylation and a decreased transcription of Toll-Like Receptor 2 (TLR 2) in periodontitis tissues was reported [15] and the methylation of E-Cadherin and Cyclooxygenase-2 (COX-2) was investigated in patients with CP and breast cancer [16]. These findings confirm the present view that chronic inflammation and cancer may have a similar epigenetic pattern and that DNA methylation may be a link between inflammation and cancer [17]. The hypermethylation of the Prostaglandin-Endoperoxide Synthase 2 (PTGS 2) promoter in inflamed periodontal tissue was associated with lower levels of PTGS 2 transcription that influence gene expression [18]. Interestingly, a CpG site close to a nuclear factor (NF)-kB transcription factor-binding site was highly methylated, suggesting that methylation status may exclude binding of this transcription factor. This is in association with other studies on methylation of cytokine promoter regions in which the CpG sites analyzed were in close proximity to binding sites for transcription factors NF-kB and Specificity protein 1 (Sp1) [13,19]. These two factors are known to be strong regulators of gene expression of the immune response.
Histone Modification: The basic unit of chromatin is the nucleosome. It consists of a DNA segment and eight core histones. The regulation of gene transcription through the post-translational modification of core histones either condenses or relaxes the chromatin [8]. The modification of histones takes place mostly at the N-terminal tails of the protein [20]. Acetylation of the core histones results in an ‘open’ chromatin conformation that facilitates transcription. Deacetylation of histones removes the acetyl groups, causing the chromatin to become more condensed and inhibit gene transcription [Table/Fig-4]. On the contrary, methylation can either result in an activated or a repressed chromatin state [5].
Acetylation and aeacetylation of histones [5].
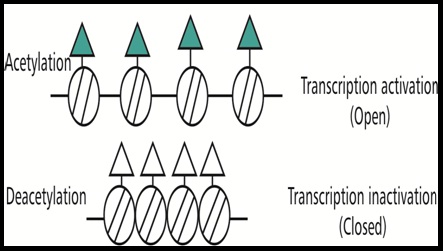
It was established that the process of DNA methylation and histone modification are linked when the CpG island of a promoter becomes methylated, methyl-CpG binding proteins bind to these methylated CpGs and recruit histone deacetylases. A strong electrostatic interaction occurs between the positively charged acetylated histone residues and the negatively charged DNA causing the deacetylation of histone proteins. Then the gene expression is repressed creating a condensed nucleosome particle. Very few studies are available on histone modifications in periodontitis. One study in experimental periodontitis has showed the importance of maintaining histone acetylation of genes related to osteoclastogenesis for preventing bone loss. Treatment with Histone Deacetylase Inhibitor (HDACi) resulted in increased bone levels [21]. The differentiation and mineralization of dental pulp stem cells have been reported through histone modification which further benefit in pulp repair and regeneration [22].
Clinical Strategy Targeting Epigenetic Modifications: A confined research has been done using HDAC or DNA methylation inhibitors as treatment for periodontitis or oral inflammation. There are various reports on the use of epigenetic inhibitors in cancer research.
In an experiment, periodontitis, treatment with HDAC inhibitor (1179.4b) (Class I and II HDACi) showed reduced alveolar bone loss in mice with P. gingivalis – induced periodontitis compared with untreated mice [23]. Imai K et al., reported that P. gingivalis produces butyric acid which inhibits HDACs and increases histone acetylation. This in turn was shown to reactivate Epstein-Barr Virus (EBV) as well as human immunodeficiency virus 1, suggesting that periodontal disease may contribute to EBV-related diseases [24].
The Bromodomain and Extraterminal Domain (BET) proteins are the epigenetic regulatory proteins. They scan the acetylated histone tails and convert transcription complexes to regulate gene transcription. A recent study by Meng S et al., (2014) showed that the BET inhibitor JQ1 was found to inhibit both an inflammatory response and alveolar bone loss in experimental periodontitis [25].
Future Scope of Epigenetics: In India, there are various genomic researches made in the field of medicine. Whereas, in dentistry the role of genetics to rule out the disease or aiming to provide treatment based on genetic variations is not much emphasized. Most researches in periodontology are focused on molecular analysis that primarily determines the aetiology, pathogenesis and prognosis of the disease. To implement in a genetic research is cumbersome, but the wide use of genome wide analysis in future can lead to a preventional regime.
Conclusion
The knowledge on the role of epigenetics in development of periodontal diseases is still constrained though the prevalence of the disease is higher in India. A few studies mentioned in this review highlights that both DNA methylation and histone modifications occur in the oral mucosa in response to bacteria and the inflammatory processes. Hence, identifying the genetic factors and epigenetic variations in periodontitis will be useful in developing innovative therapeutic interventions.
Ethical approval: This article does not contain any studies with human participants or animals performed by any of the authors.
[1]. Socransky SS, Haffajee AD, Periodontal microbial ecologyPeriodontol 2000 2005 38:135-87. [Google Scholar]
[2]. Lindroth AM, Parl YJ, Epigenetic biomarkers: A step forward for understanding periodontitisJ Periodontal Implant Sci 2013 43:111-20. [Google Scholar]
[3]. Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A, An operational definition of epigeneticsGenes Dev 2009 23(7):781-83. [Google Scholar]
[4]. Barros SP, Offenbacher S, Epigenetics: Connecting environment and genotype to phenotype and diseaseJ Dent Res 2009 88:400-08. [Google Scholar]
[5]. Bayarsaihan D, Epigenetic mechanisms in inflammationJ Dent Res 2011 90:9-17. [Google Scholar]
[6]. Kaikkonen MU, Lam MT, Glass CK, Non-coding RNAs as regulators of gene expression and epigeneticsCardiovasc Res 2011 90:430-40. [Google Scholar]
[7]. Wilson AG, Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseasesJ Periodontol 2008 79(8):1514-19. [Google Scholar]
[8]. Ji-Yun Seo, Yoon-Jung Park, Young-Ah Yi, Ji-Yun Hwang, In-Bog Lee, Byeong-Hoon Cho, Epigenetics: General characteristics and implications for oral healthRestor Dent Endod 2015 40(1):14-22. [Google Scholar]
[9]. Jones PA, Liang G, Rethinking how DNA methylation patterns are maintainedNat Rev Genet 2009 10:805-11. [Google Scholar]
[10]. Barros SP, Offenbacher S, Modifiable risk factors in periodontal disease: Epigenetic regulation of gene expression in the inflammatory responsePeriodontol 2000 2014 64:95-110. [Google Scholar]
[11]. Andia DC, de Oliveira NF, Casarin RC, Casati MZ, Line SR, de Souza AP, DNA methylation status of the IL-8 gene promoter in aggressive periodontitisJ Periodontol 2010 81:1336-41. [Google Scholar]
[12]. Zhang S, Crivello A, Offenbacher S, Moretti A, Paquette DW, Barros SP, Interferon-gamma promoter hypomethylation and increased expression in chronic periodontitisJ Clin Periodontol 2010 37:953-61. [Google Scholar]
[13]. Zhang S, Barros SP, Moretti AJ, Epigenetic regulation of TNFA expression in periodontal diseaseJ Periodontol 2013 84:1606-16. [Google Scholar]
[14]. Stefani FA, Viana MB, Dupim AC, Expression, polymorphism and methylation pattern of interleukin-6 in periodontal tissuesImmunobiology 2013 218:1012-17. [Google Scholar]
[15]. de Faria Amormino SA, Arao TC, Saraiva AM, Gomez RS, Dutra WO, da Costa JE, Hypermethylation and low transcription of TLR2 gene in chronic periodontitisHuman Immunobiol 2013 74:1231-36. [Google Scholar]
[16]. Loo WT, Jin L, Cheung MN, Wang M, Chow LW, Epigenetic change in E-cadherin and COX-2 to predict chronic periodontitisJ Transl Med 2010 8:110-15. [Google Scholar]
[17]. Kundu JK, Surh YJ, Inflammation: Gearing the journey to cancerMutat Res 2008 659:15-30. [Google Scholar]
[18]. Zhang S, Barros SP, Niculescu MD, Moretti AJ, Preisser JS, Offenbacher S, Alteration of PTGS2 promoter methylation in chronic periodontitisJ Dent Res 2010 89:133-37. [Google Scholar]
[19]. Larsson L, Thorbert-Mros S, Rymo L, Berglundh T, Influence of epigenetic modifications of the interleukin-10 promoter on IL10 gene expressionEur J Oral Sci 2012 120:14-20. [Google Scholar]
[20]. Fuchs J, Demidov D, Houben A, Schubert I, Chromosomal histone modification patterns-from conservation to diversityTrends Plant Sci 2006 11:199-208. [Google Scholar]
[21]. Valinluck V, Sowers LC, Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1Cancer Res 2007 67:946-50. [Google Scholar]
[22]. Duncan HF, Smith AJ, Fleming GJ, Cooper PR, Histone deacetylase inhibitors epigenetically promote reparative events in primary dental pulp cellsExp Cell Res 2013 319:1534-43. [Google Scholar]
[23]. Cantley MD, Bartold PM, Marino V, Fairlie DP, Le GT, Lucke AJ, Histone deacetylase inhibitors and periodontal bone lossJ Periodontal Res 2011 46:697-703. [Google Scholar]
[24]. Imai K, Ochiai K, Okamoto T, Reactivation of latent HIV-1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modificationJ Immunol 2009 182:3688-95. [Google Scholar]
[25]. Meng S, Zhang L, Tang Y, Tu Q, Zheng L, Yu L, BET inhibitor JQ1 blocks inflammation and bone destructionJ Dent Res 2014 93:657-62. [Google Scholar]