Sperm motility is essential factor of fertile men. During fertilization, sperm cells require large amount of energy for their movement of flagella and active functioning. Nearly, 100 mitochondria are present in the midpiece of every mature human spermatozoon to provide energy quickly and effectively for sperm motility [1]. The oxidative phosphorylation of mitochondria generates energy in the form of ATP for flagellar movement of spermatozoa. In mitochondria, Reactive Oxygen Species (ROS) is generated during oxidative phosphorylation and increase the risk of Mitochondrial (mtDNA) damage [2]. The oxidative phosphorylation comprises a series of protein complexes that are encoded by both nuclear genes and mitochondrial genes [3]. Mitochondria contain their own genomic DNA and express independently in matrix of mitochondria. It contains 16569 base pairs that are categorised in 13 genes of respiratory chain complex subunits, along with the 22 tRNAs and 2 rRNAs (12S and 16S) involved in protein synthesis [4]. The mtDNA replicates rapidly by D-loop mechanism without proof-reading and DNA repair mechanisms. So, it enhances mutation rate 10-100 times higher than that of nuclear DNA [5]. Furthermore, sperm cells are susceptible to damage from oxidants because they lack endogenous antioxidants activity and mtDNA is attached to the mitochondrial inner membrane where ROS are continuously generated as byproducts of electron transport chain [6]. Higher frequencies of mtDNA deletions/ mutation have been identified in mitochondrial genome of affected tissues of patients and mitochondrial diseases [5].
The large scale “common” deletions of 4977 bp is the most identified cause in affected tissue of about 40% of patients with mitochondrial myopathy and related disorder [5]. The increased number of various large scale deletions such as “common” 4977-bp, 7436-bp and 7345-bp are well recognized to be associated with aging in various human tissues [7]. Kao et al., first correlated the association of the 4977-bp deletion of mtDNA with low motility of the human spermatozoa [8]. In our previous study, we also found correlation of the 4977-bp deletions of mtDNA with decrease motility of the human spermatozoa and infertility [9]. Several studies have also shown that multiple mtDNA deletions are strongly associated with defective sperm function and diminish fertility in men and these mutations cause infertility by affecting sperm motility [10,11]. On the other hand, low level of large scale deletions has been identified in human sperm mtDNA except 4977-bp large-scale deletions and not found a direct correlation between large-scale mtDNA deletions and male infertility [12]. Therefore, we investigated the correlation between large scale 7436-bp deletions in sperm mtDNA and non-motility of sperm in asthenozoospermia and Oligoasthenoteratozoospermia (OAT) infertile men.
Materials and Methods
Collection and characterisation of Human Spermatozoa
Semen samples were collected from proven 70 asthenozoospermic and 40 OAT infertile men who attended our institutional reproductive biology laboratory of Department of Physiology, MGIMS, Sevagram. Recruitment of asthenozoospermia, OAT infertile men and 50 normozoospermic fertile men taken as control were done in accordance with WHO 2010 guideline [13]. Semen samples were collected by masturbation in sterile container after 3-4 days of abstinence and kept at 37oC for 30 min, allowed to liquefy. Semen analysis was done according to WHO 2010 guideline including motility, characteristic, concentration and morphology [13]. Asthenozoospermia sample was considered if spermatozoa with <20% normal motility and OAT was consider with <20million/ml, <20% motility and <15% normal morphology of spermatozoa. Normozoospermic semen sample was taken with count >20 million/ml, >20% motility and >15% normal morphology of spermatozoa. All subjects were aged <40 years and were included after physical examination and history taking. Infertile men with varicocele, cryptorchidism, testicular-pathy, vasectomy, long term medical history with medication, cigarette smoking and alcohol intake were excluded from our study. Informed consent form was obtained from each subject and study was approved by Mahatma Gandhi Institute of Medical Sciences Institutional Ethical Committee.
Fractionation and Preparation of sperm sample
Fractionation of human spermatozoa was done on the basis of percentage of motility by using two steps (40% and 80%) discontinuous percoll gradient technique. 40% and 80% percoll solution were prepared by mixing 100% percoll solution with Ham’s F10 medium of different volume. Fresh semen samples were washed with 1X Phosphate Buffer Saline (PBS) buffer and 500μl sperm samples was layered on top of percoll gradient and incubated at 37°C for 90 min in 5% CO2 incubator [9]. After incubation, fractionated spermatozoa were washed two times with 1X PBS buffer solution to remove percoll and prepared an aliquot of 2-3 million sperm per ml. To avoid contamination of other cells, sperm sample was treated with 50 mM Tris-Cl buffer (pH 6.8) at 6°C for 30 min prior to DNA extraction. After incubation, sperm pellet were collected by centrifugation.
Sperm mtDNA Extraction
The total DNA (nuclear DNA and mtDNA) of human spermatozoa was extracted following our standardised laboratory protocol [9]. The sperm pellet was resuspended in 0.3ml lysis buffer with proteinase-K and 10% SDS and incubated at 55oC for 2 hours. After digestion, aqueous solution was extracted with phenol:chloroform and DNA was precipitated with 3M sodium acetate and chilled absolute ethanol. After washing with 75% ethanol and dissolve in 1X TE buffer (pH 8), then DNA was ready for PCR.
Long range amplification and primer shift PCR
The long range PCR amplification of targeted sequences of mtDNA was performed with 100-150ng of template mtDNA in 50μL of total reaction volume, containing 200 mM of each dNTP, 1mM of light chain and heavy chain primers [Table/Fig-1], 2U of herculase DNA polymerase, 50mM KCL, 2mM Mgcl2, 25mM Tris(hydromethyl) methyl-3-aminopropanesulphonic acid (Agilent Technologies, USA), 1mM – mercaptoethanol, and 10mM Tris-HCL (pH 8.5). PCR was carried out for 25 cycles in two steps in first 13 cycles using the denaturation at 94°C for 1 min, annealing at 55–58°C for 1min, and primer extension at 72°C for 8 min and remaining 17 cycles denaturation and annealing condition were same but primer extension at 68°C for increasing 10 seconds to each reactions [8,9]. The amplified PCR products were then separated on a 1% ethidium bromide containing agarose gel for 1.5 hour and observed on gel documentation system (Uvi-Tec, UK). The presence or absence mtDNA was determined using two primer pairs of Mitochondria Light Chain (MTL)-2, Mitochondria Heavy Chain (MTH)-4, same primer sets also used for confirmation of deletion finally. The primer sets MTL2-MTH4, MTL2-MTH5, MTL2-MTH6 were used for long range PCR amplification and primer shift technique was followed for the identification of existence of 7436 bp large scale deletions of mtDNA in asthenozoospermic and OAT semen samples (PCR products amplified from each of the primer pairs are shown in [Table/Fig-2]).
Sequences of primers used for primer shift technique to amplify mtDNA for detection of 7436-bp deletion (Kao et al., 1998)
| Primers | Primers Sequences | NucleotidePositions (bp) |
|---|
| MTL2(Mitochondria light chain 2) | 5’-GCCCGTATTTACCCTATAGC-3’ | 8251-8270 |
| MTH4(Mitochondria heavy chain 4) | 5’-TGTAGCCGTTGAGTTGTGGT-3’ | 10149-10129 |
| MTH5(Mitochondria heavy chain 5) | 5’-AGGAACCAGATGTCGGATAC-3’ | 16509-16490 |
| MTH6(Mitochondria heavy chain 6) | 5’-CGAGGAGAGTAGCACTCTTG-3’ | 16450-16431 |
Nucleotide primer pairs used for the analysis of the 7436-bp deletions in the mitochondrial DNA (mtDNA) of human sperm.
| Primer Pair | Amplified region | PCR product length (normal mtDNA) | PCR product length (in deleted mtDNA) |
|---|
| MTL2-MTH4 | 8251-10149 | 1898-bp | — |
| MTL2-MTH5 | 8251-16509 | 8258-bp | 822-bp |
| MTL2-MTH6 | 8251-16450 | 8199-bp | 763-bp |
MTL - Mitochondria Light Chain; MTH - Mitochondria Heavy Chain
Results
Results of percoll fractionation of sperms of patients and controls showed that low density 40% percoll fractions had contained non-motile or poor motile spermatozoa and high density 80% percoll fractions had highly motile spermatozoa.
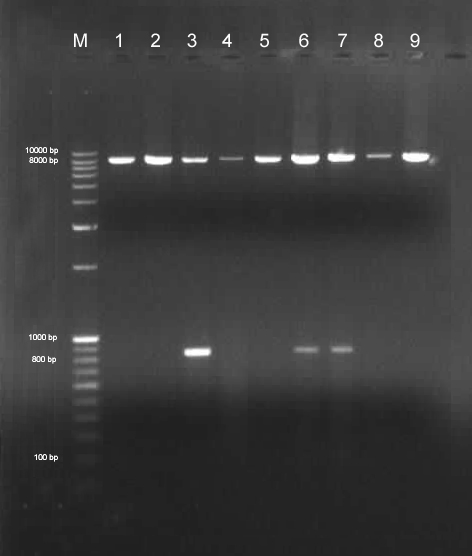
The analysis of large scale deletions of 7436 bp mtDNA revealed that spermatozoa from 40% and 80% percoll fractions had 8 (7.2%) and 3(2.7%) mtDNA deletion respectively in infertile males using MTL2-MTH5 primer set. Out of one hundred and ten patients, 8 (7.2%) patients had showed deletions of 7436-bp (np 8637 bp to np 16073 bp). Of the eight patients showing sperm mtDNA deletions, six were asthenozoospermic (6/70; 8.5%) infertile men and two were OAT (2/40; 5%) infertile men [Table/Fig-3]. The full length PCR product of 8258-bp was observed from the wild-type mtDNA and approximately 763-bp to 822-bp bands were generated from the 7436-bp deleted sperm mtDNA. The primer shift method clearly had shown the presence of 7436-bp deletion in sperm mtDNA. The primers MTL2-MTH5 and MTL2-MTH6 were used for primer shift amplification of mtDNA and with the following primers we obtained the PCR products of 822-bp and 763-bp respectively from the 7436-bp deleted sperm mtDNA [Table/Fig-4]. Fifty samples of control fertile normozoospermic men spermatozoa were also analysed for 7436-bp mtDNA deletions from 40% and 80% percoll fractions and no such deletions were observed in both fractions.
Proportion of the 7436-bp deletion in 40%, 80% Percoll fractions of asthenozoospermia (asthe), oligoasthenoteratozoospermia (OAT) and normal fertile males.
| SampleClassification | No. of Sample | Percoll Fractions | Sample-wise % of subjects with Deletions(in 40% Percoll) |
|---|
| 40% | 80% |
|---|
| Subject-with deletion | Subject-with deletion |
|---|
| Asthe | 70 | 6 | 2 | 6/70 (8.5%) |
| OAT | 40 | 2 | 1 | 2/40 (5%) |
| Total | 110 | 8 (7.2%) | 3 (2.7%) | 8/110 (7.2%) |
| Normal | 50 | 0 | 0 | 0 |
Detection of mtDNA molecules with the 7436-bp deletions by polymerase chain reaction (PCR) method. Lane M: marker 100bp-10kb and in lane 1: control DNA, by using primer pair MTL2-MTH5 we obtained the deleted and wild type mtDNA in lane 3, 6, 7. The PCR product size of the nucleotides flanking the deleted part is 822-bp and in lane 2, 4, 5, 8, 9 shown normal 8258-bp PCR product.
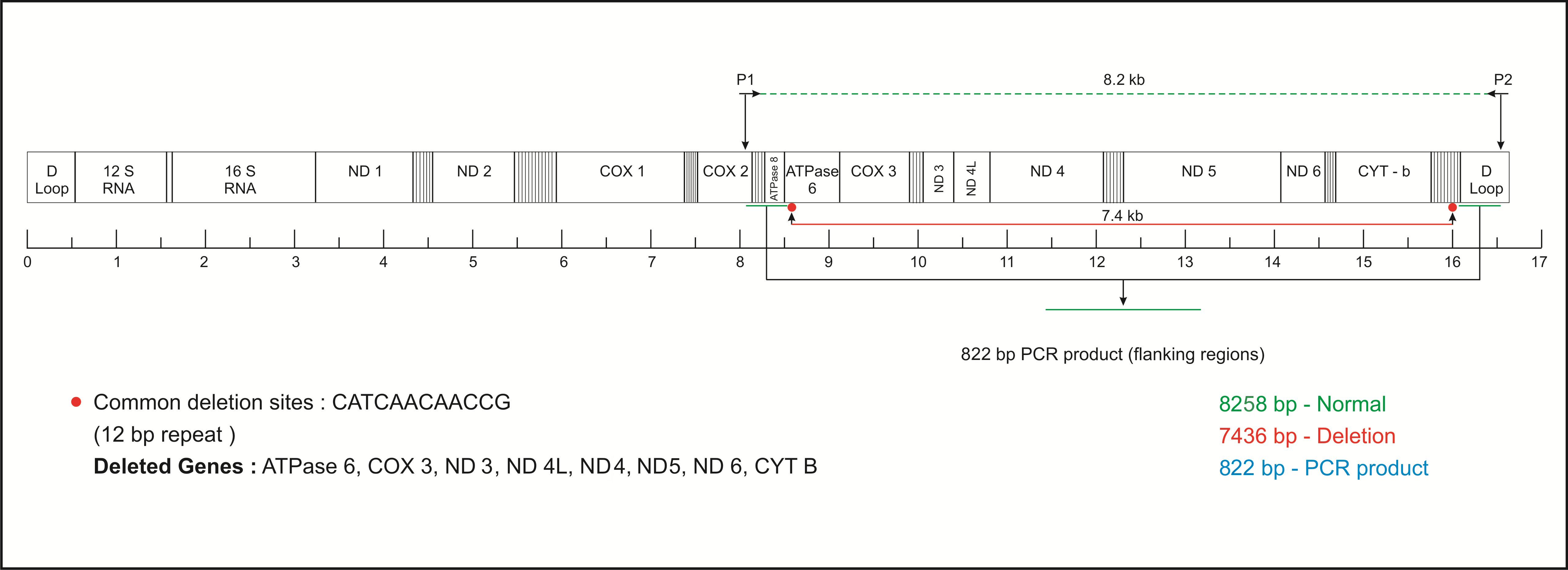
Upon close examination of the deletions sites or breakpoints and 7436-bp deletion, we found two "12-bp direct repeats" in the junction sites from np 8637 to 8648-bp and from np 16073 to 16084-bp in the mtDNA. Sequencing result of PCR products indicated that maximum breakpoints for 7436-bp deletions were present in the flanking regions of deleted segments. The break-points were present around the direct repeat of 12-bp (CATCAACAACCG) but rarely within the direct repeats. Thus, they generate different types of PCR products. The nucleotide sequences flanking the breakpoints of the deletions of 7436-bp of mtDNA in human spermatozoa are depicted in [Table/Fig-5].
A scheme illustrating the strategy used for the confirmation of large scale 7436-bp deletion of mtDNA, 12-bp repeat common deletions sites and deleted gene on mtDNA.
Discussion
Sperm motility is an important determinant of male fertility and mitochondria is an essential cell organelle to generate energy for sperm motility [14]. Non-motile spermatozoa are unable to move through the mucus layer of cervix to reach the sites of fertilization. Therefore, active motility of spermatozoa is essential for successful fertilization [15]. Several factors such as genital infections, defects in flagellar mechanism, maturation defect in the epididymis and abnormality in oxidative phosphorylation process can decline sperm motility and abnormal morphology of spermatozoa in asthenozoospermia and OAT infertile men [16]. Various types of sperm mitochondrial DNA deletions/ mutations have been reported in asthenozoospermia and OAT infertile men [8,9].
Analysis of our results revealed that 8.5% asthenozoospermic patients and 5% OAT patients had showed 7436-bp sperm mtDNA deletions and overall 7.2% patients had showed deletions of 7436-bp sperm mtDNA in infertile men when the sperm samples were collected from 40% percoll fractions. But, the samples from fertile normozoospermic subjects (control) did not show any such deletions. Thus, comparing the results from the subjects and controls there is a definite indication that 7436-bp deleted mtDNA in asthenozoospermia and OAT patients play a major role for sperm motility. Sequencing of PCR products indicated the break-point of large scale deletions and confirmed that the break-points were present in or around the two direct 12-bp repeats of mtDNA. In mtDNA, direct repeats are vulnerable sites for large-scale DNA deletions [17]. Studies have demonstrated that the 7436-bp large scale deletions were present in mt DNA of somatic tissues along with the mtDNA of infertile male spermatozoa [18]. Large scale 7436-bp deletions in non-motile or poor motile spermatozoa are less frequent than the "common" 4977-bp large scale deletions in mtDNA [8]. Different mechanisms have been proposed for the large scale deletions involving direct repeats present in mitochondrial genome [19,20]. Fukai and Moraes advocated that large scale deletions occurred by slipped-mispairing, illegitimate recombination, oxidative stress created by free radicals, and topoisomerase or DNA recombinase-mediated DNA breaks but still the actual mechanism of mtDNA deletions is unclear [20].
The large-scale deletions result in complete removal or truncation of some structural genes such as ATPase 6, COX-3, ND3, ND4L, ND4, ND5, ND6, CytB and eight tRNA genes of mtDNA [21]. The defective protein subunits formed by deleted or mutated mtDNA when assembled with nuclear encoded subunits lead to abnormality in respiratory enzymes [22]. Spermatozoa containing defective mtDNA not only produce ATP less efficiently, but also generate more ROS and free radicals, which may further damage mitochondria and mtDNA leading to an ultimate energy crisis and decline of motility and fertility [23]. In mitochondria, oxidative phosphorylation processes continuously produce exogenous, and endogenous free radicals with other ROS and it increases risk of mtDNA damage [21]. Generation of excess ROS and low anti-oxidant levels in the semen might cause mutation/deletions in mtDNA and impair the fertilizing capacity of spermatozoa [23]. During spermatogenesis, mtDNA deletions/mutations may occur and accumulate in the spermatids and ultimately damage respiratory function and decreases sperm motility [24]. In a study of, Kumar et al., they have reported mutations in ATPase6, ATPase8, ND2, ND3, ND4, ND5, COX3 mitochondrial genes in non-motile spermatozoa of the OAT infertile men [25]. Kao et al., showed correlation of increased ROS mediated oxidative stress with 4977-bp and 7436-bp large-scale deletions of mtDNA in non-motile spermatozoa [21]. In our previous study we also noted high frequencies of "common" large scale 4977-bp sperm mtDNA deletions in non-motile spermatozoa of asthenozoospermic and oligoasthenoteratozoospermic subjects [9]. Abasalt et al., recorded positive correlation between large scale (4.8, 7.4-kb) mtDNA deletions and abnormal human sperm motility in asthenozoospermia and OAT infertile men [26].
Results of the earlier studies [21,26] and also our previous study [9] confirmed the role of 4977-bp "common" deletions in sperm mtDNA of infertile subjects with asthenozoospermia and OAT. Some reports confirmed the association of large scale 7436-bp sperm mtDNA deletions in asthenozoospermia and OAT patients [8,12,18]. But, the frequencies of 7436-bp deletions are less than that of 4977-bp "common" deletions [26]. In the present study, we observed 7.5% deletions of 7436-bp mtDNA in patients, but there were no such deletions in fertile normozoospermic control subjects. Thus, the results indicate that large scale 7436-bp sperm mtDNA deletions play a vital role in infertile men with asthenozoospermic and oligoasthenoteratozoospermic.
Limitation
We have identified large scale mtDNA deletions by long-range PCR technique. Now, site specific deletions or mutations are needed to be studied by using more number of primers and sequencing of mtDNA which didn’t show any deletions in asthenozoospermia and OAT subjects. These parameters are beyond our present study protocol. In future study, these limitation need to be addressed.
Conclusion
Non-motile abnormal sperms of asthenozoospermia and OAT cases had the mtDNA deletions of 7436-bp DNA segment which actually contains the genes responsible for oxidative phosphorylation. Normal motile sperm did not show such deletions. Oxidative phosphorylation is the source of energy for the sperm-motility and the deletions are responsible for poor sperm motility. Our result indicates that the deletions of large scale 7436-bp in mtDNA were responsible for poor sperm motility and had been associated with sperm dysfunction and male infertility.