Materials and Methods
The study was conducted between 2009-2011 in department of Microbiology, Jawaharlal Nehru Medical College, KLE Dr. Prabhakar Kore Charitable Hospital & Medical Research Centre and Regional Medical Research Center (Indian Council of Medical Research; RMRC-ICMR) Belagavi. Institutional ethical clearance (IEC) was obtained before commencement of the study. 300 stool samples from acute diarrhoea patients reporting to our tertiary care hospital were included. There were 14 patients of < 1 year, 68 patients in 1-4 age group, 75 patients in 5-9 years of age group and 143 patients >10 years of age. Patients already on antibiotics and those with provisional diagnosis of diarrhoea due to metabolic causes were excluded from the study. Detailed history of each patient was recorded in pre-tested diarrhoea case reporting form and stool samples were collected in transparent, clean, sterile, screw capped 20mL capacity vials. These samples were subjected to microscopy for detection of ova/cysts of protozoan parasites, helminths and culture on Mac Conkey and blood agar plates for isolation and identification of common diarrhoeagenic bacterial pathogens (Vibrio, Shigella species, Salmonella species and E.coli) by standard microbiological methods. Rapid lysates of E.coli isolates from those stool samples which yielded only E.coli on Mac Conkey and Blood agar plates were subjected to multiplex PCR for determining presence of stx and eae gene. These E.coli were also submitted for serotyping to National Salmonella & Escherichia Centre/Research and Development Laboratory, Central Research Institute, Kasauli (Himanchal Pradesh), India which provided only the serotypes and their affiliation was determined through published scientific literature [11–13].
For PCR, DNA was extracted using Cetyl Trimethyl Ammonium Bromide (CTAB) method [14] and quality of the DNA was checked on agarose gel electrophoresis by viewing 1μl of the stained DNA under UV light in a Gel-documentation system (Alphaimager, USA). The quantity of DNA was estimated spectrophotometrically by using a Nanodrop spectrophotometer (JH Biosystems, USA). A 260/280 nm for detection of any impurity in DNA samples. The stx gene was detected in PCR for identification of shiga toxin producing E.coli using, primers CAGTTAATGTGGTTGCGAAG and CTGCTAATAGTTCTGCGCATC amplifying an 895 bp segment of the DNA as described in WHO Manual for Laboratory Investigations of acute enteric infections [15]. For eae the primer sequences used were AAACAGGTGAAACTGTTGCC and CTCTGCAGATTAACCCTCTGC as described by Yu J et al., [16]. The PCR conditions are enumerated in [Table/Fig-1]. For determining the genetic diversity of the STEC isolates, RAPD fingerprinting assay was used [17,18] in which PCR was performed at least three times by different technicians in 50μl reaction volumes mixture containing 50ng DNA, 0.3μm primer PB1 (5’-GCG CTG GCT CAG-3’). PCR for stx, eae and RAPD were performed in BioRad iCycler thermal cycler (BioRad, USA) and all reaction products were electrophoresed on 20cm long 1% agarose gels, stained with 0.5μg/ml GelRed (Biotium, USA), and photographed under ultraviolet light using gel documentation system (AlphaImager, USA). For RAPD fingerprinting DNA profiles of individual lanes were matched with each other and dendrograms of distance based on similarity coefficient were generated with the Unweighted Pair Group Method using Arithmetic averages (UPGMA) for clustering at 5% confidence level [17,18].
PCR cycling conditions for stx, eae and RAPD.
| PCR for | PCR reaction mixture | Cycling condition |
|---|
| stx and eae | 10X PCR Buffer with MgCl2: 2.5μldNTP mix (2.5mM each): 2.0μlForward Primer (stx) 10μM : 1.0μlReverse Primer (stx) 10μM: 1.0μlForward Primer (eae) 10μM: 1.0μlReverse Primer (eae) 10μM: 1.0μlTemplate DNA (lysate): 2.5μlSterile water: 13.8μlTaq polymerase (5U/ μl): 0.2μlTotal volume: 25.0μl | Denaturation 94°C for 1 minAnnealing 55°C for 1.5 minExtension 72°C for 1.5 minFinal extension 72°C for 7 minTotal number of cycles required:30 |
| RAPD | 50μl reaction volumes mixturecontaining 50 ng DNA, 0.3μmprimer PB1 (5’-GCG CTGGCT CAG-3’),250μm of each dNTP, 3mMMgCl2, 0.5U Taq DNApolymerase, in 10mM HCland 50 mM KCl. | One cycle of 7 min at 94 °C,1 min at 40 °C and 1 minat 72 °C. Four cycles of 1min at 94 °C, 1 min at 40 °C,and 1 min at 72°C. Twentyfour cycles of 1 min at 94 °C,1 min at 55 °C, and 1 minat 72 °C; and the final cycle of1 min at 94 °C, 1 min at 55 °Cand 7 min at 72 °C |
Results
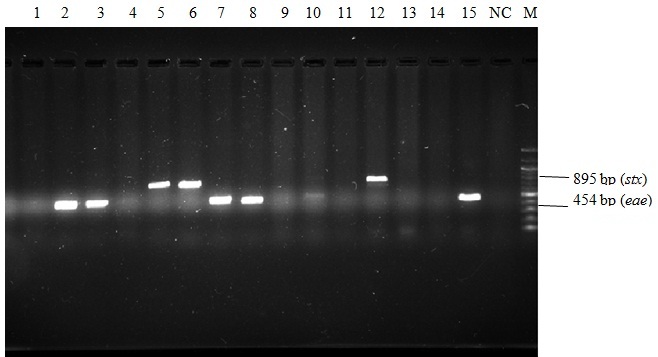
Out of 300 samples, 102 samples yielded one of the common diarrhoeagenic pathogens viz. Vibrio cholera/Salmonella species/Shigella species/protozoan parasites/ significant amount of Candida with pseudohyphae either by microscopy or culture [Table/Fig-2]. Remaining 198 cases yielded pure growth of E.coli on blood agar and Mac Conkey agar culture plates. PCR detected eae (responsible for attachment and effacement) in 118 isolates, stx (coding for shiga toxin) in 55 isolates and presence of stx and eae together in 46 (23.2%) out of 198 culture isolates [Table/Fig-3]. On the contrary, serotyping identified only three isolates of E.coli as STEC and none of these three isolates belonged to serotype O157:H7. These isolates belonged to O59, O60 and O69.
Distribution of Protozoan / Helminthic parasites, Salmonella spp, Shigella spp, Vibrio cholera and Candida spp. in sporadic cases of acute diarrhoea amongst various age groups (n= 300).
| Pathogens | Age in years(Number ofcases in thatage group) | Numberof samples |
|---|
| <1(14) | 1-4(68) | 5-9(75) | 10+(143) | Positive(%) | Negative |
|---|
| ProtozoanParasites(Trophozoites/ Cysts) | 0 | 0 | 1 | 7 (4.8%) | 8 (2.6) | 292 |
| Helminthic Ova | 0 | 0 | 8 (10.7%) | 33 (23.1%) | 41 (13.6) | 259 |
| Vibrio cholera | 0 | 0 | 0 | 6(4.1%) | 6 (2) | 294 |
| Salmonella spp. | 0 | 0 | 0 | 19 (13.2%) | 19 (6.3) | 281 |
| Shigella spp. | 0 | 0 | 3 (4%) | 8 (5.6%) | 11 (3.6) | 289 |
| Candida spp. | 0 | 0 | 10 (13.3%) | 7 (4.8%) | 17 (6) | 283 |
| Total | 0 | 0 | 22 (29.3%) | 80 (55.9%) | 102 (34%) | |
Multiplex PCR for stx (895 bp) and eae (454 bp) genes.
• Lanes 5, 6 & 12 showing DNA bands positive for stx gene.
• Lanes 2, 3, 7, 8 & 15 showing DNA bands positive for eae gene.
• Lane NC denotes negative control.
• M denotes marker DNA 100 bp ladder (covering 100 bp to 1200 bp with more intense 500 and 100 bp sizes).
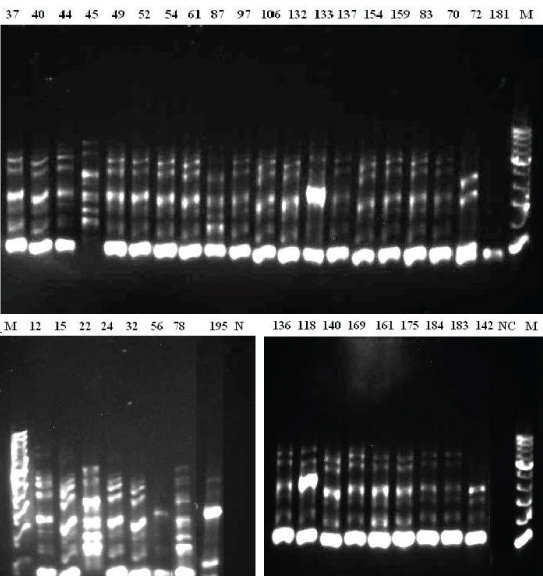
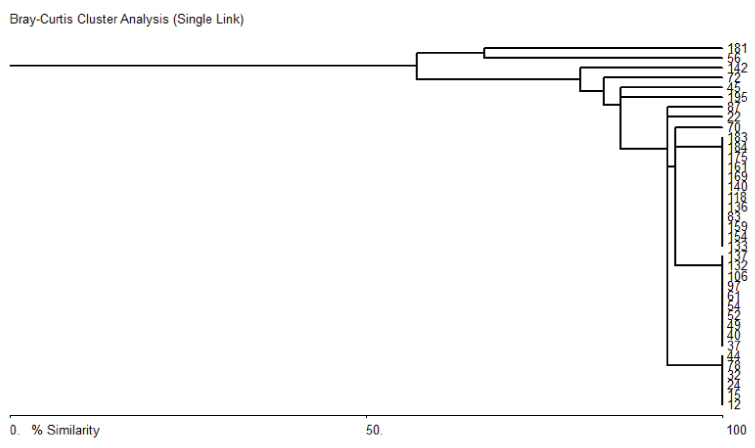
Eleven clones of STEC were identified in 46 STEC isolates by RAPD fingerprinting [Table/Fig-4]. RAPD fingerprinting assay showed that most of the STEC isolates were genetically close to each other. There were three large groups consisting of 12, 10 and 6 isolates, members of which were genetically identical (100%). Between these groups, the isolates shared more than 90% similarity. There were another 3 isolates outside these groups which shared more than 90% similarity with these groups. There were 6 more isolates that did not cluster with others and shared 55 to 75% similarity to the others [Table/Fig-5].
Fig. 2: RAPD fingerprinting for STEC isolates NC: Negative control. M: 100 bp DNA ladder.
Dendrogram of STEC isolates.
11 different clones of STEC were observed. Interestingly while 3 particular strains were common with 12, 10 and 6 isolates belonging to these groups respectively, there were as many as 8 clones those where represented only by one isolate. This means that there was high diversity in the isolated STEC stains suggestion that cases were likely from multiple sources
11 different clones of STEC were observed. Interestingly while 3 particular strains were common with 12, 10 and 6 isolates belonging to these groups respectively, there were as many as 8 clones those where represented only by one isolate. This means that there was high diversity in the isolated STEC stains suggestion that cases were likely from multiple sources.
Discussion
It is to be acknowledged that non- O157:H7 E.coli have potential to cause outbreaks of severe illness and these infections can transcend beyond the national boundaries, which was evident from last outbreak involving Germany, USA and France [10].
Detection of 46 (23.2%) cases of STEC mediated diarrhoea as determined by presence of stx and eae by PCR in 198 cases of acute diarrhoea presumably caused by E.coli brings about two very important findings viz., significant presence of STEC in this geographical area and these isolates do not belong to O157:H7 serotype. Outbreaks in developed countries due to STEC, even though involving less number of people, draw very significant attention and publicity because very few cases due to food contamination and consequent faecal-oral transmission are expected due to good hygienic practices in these countries. In developing countries the scenario is very different and water borne infections, primarily due to faecal contamination are very common. Surprisingly STEC other than O157:H7 serotype is not actively sought in either of the epidemiological settings even though non-O157:H7 infections have been made a nationally notifiable condition since 2005 in USA [19].
The discovery of stx was made 100 years ago, much earlier than the description of cytotoxic assay in 1977 by Konowalchuk and it was recognized much later that over 100 different E.coli serotypes can express stx [20,21]. Because stx producing bacteria are associated with asymptomatic intestinal colonization as well as with life threatening HUS [1] and it is not very clear whether mere possession of genes encoding stx confer pathogenicity, we looked for the presences of intestinal adherence factors coded by eae in addition to stx to establish definitive role of stx positive E.coli isolates in causation of diarrhoeal illness even though these strains were isolated from diarrhoea cases itself. The reason for choosing intestinal adherence factors coded by eae gene along with stx for identification of STEC as the diarrhoeagenic shiga toxin producing E.coli (STEC) was based upon the fact that in conventional and gnotobiotic piglets, O157:H7 strains produce extensive A/E (attaching and effacing) lesions in the large intestine, featuring intimate adherence of the bacteria to the epithelial cells [9]. In contrast, O157:H7 strains specifically mutated in the eae gene no longer produced A/E lesions and did not appear to colonize any intestinal site [13–15]. Additional support for a role in human disease is seen with the anti-intimin immune response seen in HUS patients and, by extrapolation, the reduced virulence in volunteers challenged with an EPEC strains mutated in eae [22].
In addition to initial ham-burger associated outbreaks, sources of EHEC infections include pink ground beef, mayonnaise, uncooked radish, alfalfa sprouts, spinach which usually carry faecal contamination of cattle who are known reservoir of stx producing E.coli (mostly other than O157:H7), and even unpasteurized apple juice and fermented hard salami etc [9]. The presence of EHEC in apple juice and fermented hard salami is an ample evidence for the ability of STEC growth at much acidic pH 3.4 which is non-conducive for many pathogens.
In July 2011, there were 852 reported patients of HUS and 32 HUS associated deaths in Germany due to O104:H4 infection, the source of which was contaminated raw sprouts produced in a farm in Lower Saxony in Germany. The infection reached to USA and six confirmed cases (including one death) of same serotype were reported of which five cases had travelled to Germany in that period. During the same period, France reported cluster of E. coli O104:H4 infections among people who attended an event in Bordeaux and reported eating sprouts served at the event [10]. Aforementioned outbreak originated in country where there is very little scope for faecal contamination as opposed to developing countries with plenty of opportunities for faecal-oral contamination to occur.
As per 2008 WHO report more than 600 million people practice open defecation in India [23] which is one of the fastest growing economies and even though there are significant strides in provisions for improved water supply there always remain the possibility of faecal-oral contamination as evident from the fact that in India diarrhoea accounts for 24% to 30% of deaths in infants aged 1 to 11 months [23]. The fact that non-O157:H7 STEC was detected in 23% of stool samples collected from cases of acute diarrhea in a tertiary care hospital points to a grim situation which may acquire larger proportion anytime and given the realty of shrinking distances and travel time, can trigger outbreaks at faraway places, otherwise having good containment for these pathogens. It is within reasonable limits to infer that situation may not be different in other developing countries as well. Therefore there is an urgent need to actively test stool samples for non- O157:H7 STEC serotypes, which is not a standard practice even in all developed countries.
Stigi et al., had observed in 2010 that 37 laboratories in Washington who processed nearly half of the stool cultures in the state attempted culture only for O157:H7 and therefore could not detect non-O157:H7 STEC, though in the same study increase in cases of non- O157:H7 was reported which corresponded with increased number of laboratories attempting to test for non- O157:H7 [24].
The fact that only three isolates were identified as STEC by serotyping performed at a national reference laboratory as compared to PCR which detected 46 stx positive E.coli underlines inadequacy of serotyping for detecting all serotypes of STEC. It is to be further taken into consideration that conventional method of identifying STEC on the basis of inability to ferment sorbitol by O157:H7 will also not hold well because many of the non-O157:H7 serotypes ferment sorbitol [9]. Under such circumstances, detection of stx gene by PCR offers many advantages in diagnosing STEC infections including non-O157:H7 serotypes which, irrespective of their serotype, will harbor stx gene that can be detected by PCR.
Eleven different clones of STEC were detected by RAPD fingerprinting assay amongst the 46 STEC isolates. Although reproducible fingerprints were obtained for only 34 of the STEC isolates in RAPD assay, the results show that there is sufficient genetic diversity in the isolates pointing perhaps to infections occurring with different strains over different times and locations and that RAPD is a reasonably good technique for determining extant of genetic relatedness.
Although the use of RAPD as a technique has several limitations, it is quite reliable and easy to perform for identifying diversity and clonal relatedness for bacterial pathogens. Presence of multiple clones/genotypes indicates the possibility of multiple sources contaminating the water supply/food and existence of these organisms in various places in the region over quite a period of time. The implication of these findings will be of use for the efforts directed towards interventions to plug the breaches in the water supply to ensure access to safe water, even though it will be a difficult preposition to identify multiple points of contamination and incorporate effective interventional measures because it is very common to find in developing countries the water supply getting contaminated at more than one place. Hence in such countries the best approach is to treat water at the ‘point of use’ itself.
Three most common acronyms/ nomenclature viz., VTEC (verocytotoxigenic E.coli), STEC (shiga toxigenic E.coli / shiga-like toxin producing E.coli) and EHEC (Enterohaemorrhagic E.coli) represent one group of Escherichia coli [25–27]. VTEC denote the ability of toxin produced by this group of Escherichia coli to bring about demonstrable changes on Vero cell lines in tissue cultures. STEC indicate resemblance of this toxin to that produced by Shigelladysenteriae and EHEC is more of a clinical terminology denoting presence of haemorrhagic colitis. It is also a well-established fact that all E.coli possessing stx gene/producing shiga toxin do not cause HUS [2–4]. Hence, among these acronyms shiga-like toxin producing E.coli is more accurate and should be used in all those situations wherein stx harboring E.coli is identified but shiga toxin production is not demonstrated by using cell culture and clinical entity of HUS has not been established.
Limitation
Organisms like Campylobacter, coccidian parasites and diarrhoeagenic viruses have been excluded in the study. Even though the possibility of co-infection with protozoan parasites, and other bacterial pathogens including Vibrio, Shigella species, Salmonella species included in this study is minimal, but it is not evaluated and the same can be considered another limitation of this study.
Conclusion
About 15% of all diarrhoea cases included in the study were caused by STEC which accounted for 23% of diarrhoeagenic E.coli isolates. These isolates can assume epidemiological significance as reported in previous studies and for this reason laboratories shall actively look for them and report. Conventional culture cannot differentiate between commensal E.coli and diarrhoeagenic E.coli. Serotyping is not reliable for identification of STEC coupled with the fact the non-O157:H7 E.coli are important causative pathogen for sporadic diarrhoea as evident from this study. Detection of stx and eae genes by PCR is better modality especially when PCR are becoming available in most of the laboratories. It is imperative to understand and state that though all EHEC strains have stx gene but not all stx possessing E.coli can cause haemorrahgic colitis and HUS. Hence, we are of the opinion that use of EHEC term shall be restricted only to those E.coli strains isolated from cases of haemorrahgic colitis / HUS. For other E.coli isolated from diarrhoea cases and shown to possess stx, STEC will be more appropriate, since cell lines for demonstrating changes in Vero cell lines are not used for routine identification.