Surgical intervention may be coupled with surgical site infection (SSI) as reported in 5% of the surgical procedures [1]. SSI may extend from easily manageable state to serious life threatening condition [1]. It still remains one of the most important healthcare associated infections causing pain, prolonged hospital stay with increased expenditure, cosmetically unacceptable scars, thus, leading to the miserable condition of the patients [2]. This entire sequel could be prevented by systemic antibiotic prophylaxis as the bacteria implicated in SSIs include those, which are conceded by the patients themselves (endogenous flora), or those that might be instigated in the operating room (exogenous flora) [3]. Infection caused by microorganisms from an external source following surgery is less frequent than the endogenous one [4].
On this background the study was conducted to determine the prevalence of MRSA in SSI and also to determine antibiotic susceptibility pattern of MRSA strains that may ultimately help the infection control team to plan the preoperative antibiotic policy.
Materials and Methods
A cross-sectional study was conducted for a period of 31/2 years (July, 2009 to December, 2012) at Nil Ratan Sircar Medical College, Kolkata, West Bengal after getting approval from the Institutional Ethics Committee.
Samples were taken from the cases of SSIs, selected according to the CDC, HICPAC guidelines, 1999 [5]. Any infection involving skin and subcutaneous tissue arising around the incision within 30 days of a clean surgery is defined as SSI. Discharges from stitch abscesses, infection of an episiotomy or neonatal circumcision site, infected burn wounds, incisional wound that extend into the fascial and muscle layers and contaminated and dirty surgeries were excluded from the study.
A total of 19,359 surgical procedures were done of which 3003 culture positive SSIs were noted. The clinical samples were collected from patients of both sexes and all ages (starting from one year of age – 6 groups were made taking subsequent 15 years in each group), suspected to be suffering from surgical site infection from different specialities like Surgery, Orthopaedics, Gynaecology & Obstetrics, Urology, Paediatric Surgery and Cardiovascular surgery. Samples were also collected from patients with infected wounds who underwent minor surgical procedures like tracheostomy, venesection, peritoneal dialysis etc. of different wards like Medicine, Paediatrics, Haematology and Chest. The area around the wound was cleaned with 70% ethyl alcohol followed by normal saline and exudates were collected from the wound with a sterile inoculating loop or with a sterile swab stick soaked in normal saline or sometimes by aspirating with a sterile syringe and needle.
The samples were inoculated on Blood agar, MacConkey’s agar and Mannitol salt agar. The plates were incubated aerobically at 37oC overnight. If growth failed to appear, incubation was continued up to 48 hours. The colonies suggestive of Staphylococcus aureus were identified by standard procedures (Gram staining, catalase test, slide coagulase and tube coagulase test, phosphatase test etc.,) [16]. Tube coagulase was taken as the main criteria for identification of Staphylococcus aureus [16].
Antibiotic susceptibility testing was done by Kirby Bauer method [17] following Clinical and Laboratory Standards Institute (CLSI) guidelines [17] using commercially available cefoxitin (30μg) disc (HiMedia) and the results were compared with Staphylococcus aureus ATCC 25923 and MRSA ATCC 43300 control strains. The other antibiotic discs used were vancomycin (30 μg), oxacillin (1μg), ciprofloxacin (5μg), netilmycin (30 μg), linezolid (30μg), gentamycin (10 μg), clarithromycin (15μg), erythromycin (15 μg), levofloxacin (5μg), clindamycin (2μg), fucidin (10 μg), mupirocin (5 μg), co-trimoxazole (25 μg) and azithromycin (15 μg). All Staphylococcus aureus strains were screened for MRSA by detection of resistance to Cefoxitin disc (zone of inhibition was ≤21 mm) following the CLSI guidelines [17]. Cefoxitin resistant strains were further subjected to Slidex staph latex agglutination tests (BioMerieux) to determine the phenotypic expression of mecA gene [18,19]. PCR was done to detect mecA for genotyping [20]. DNA extraction was done on all the isolates as described in Phenol-Chloroform Method with minor modification [21,22]. The DNA fragments of 606 bp were amplified from mecA gene using specific primers, mecA F 5’AGTTGTAGTTGTCGGGTTT3’, mecA R 5’AGTGGAACGAAGGTATCATC3’ [20,23]. The conditions for PCR were denaturation at 94oC for 5 minutes, followed by 34 cycles of an initial denaturation at 94oC for 1 min, annealing at 54oC for 1.5 mins, and extension at 72oC for 1 minute and final extension step at 72oC for 10 minutes [20]. The amplification was carried out in Thermal cycler (Biometra, Germany). The amplification product (10μl) was analysed by electrophoresis on 1% agarose gel and visualized with ultraviolet light after staining with ethidium bromide [22].
Among the MRSA strains, those found to be resistant to vancomycin (30 μg) by disc diffusion test (zone of inhibition≤ 14 mm) were further resorted to E test (BioMerieux) to determine the Minimum Inhibitory Concentration (MIC) for confirmation of their resistance status [24]. Isolates with a vancomycin MIC 4 to 8 μg ml−1 were identified as Vancomycin-intermediate Staphylococcus aureus (VISA), isolates with a vancomycin MIC >16 μg ml−1 were identified as Vancomycin-resistant Staphylococcus aureus (VRSA) [17].
Statistical analysis was done using the CDC Epi Info TM 7 Stat Calc software program. Chi square test/ Fisher Exact test were applied for comparison of categorical data. p < 0.05 was considered to be statistically significant.
Results
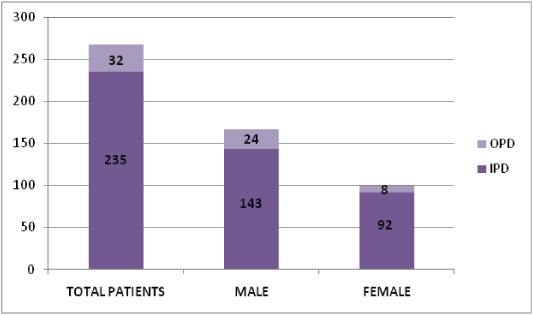
In this 3½ year study, 15.51% SSIs have been documented. SSIs were predominantly due to Staphylococcus aureus (34.93%) followed by Escherichia coli (20.34%), Klebsiella spp. (18.08%), Pseudomonas spp. (7.99%), Acinetobacter spp. (7.49%), Enterococcus spp. (4.39%), Coagulase negative Staphylococcus spp (3.16%), Proteus spp. (1.73%), Citrobacter spp. (1.29%), Enterobacter spp. (0.26%), Providencia spp. (0.16%) and Morganella spp. (0.13%). Among the 1049 Staphylococcus aureus, 267 Methicillin resistant Staphylococcus aureus (MRSA) strains were observed. Prevalence of MRSA was (267 / 1049 x 100) 25.45%. MRSA were isolated from 167 (62.54%) male patients and 100 (37.45%) female patients having surgical site infection. Inpatients and outpatients distribution of MRSA were 235 (88.01%) and 32 (11.98%) respectively [Table/Fig-1]. As the age progresses there was more incidence of MRSA infecting the wounds which were statistically significant [Table/Fig-2].
Indoor and outdoor distribution of the male and female patients of SSI with MRSA.
Age wise distribution of MRSA in SSI.
| Agedistribution | MRSA(%) SSI | Non MRSA(%) SSI | Total SSI | |
|---|
| 1-15 years | 12(3.46) | 335 (96.54) | 347 | Chi2 = 23.346with degree offreedom 5 andp-value <0.001 |
| 15-30 years | 62(7.82) | 731 (92.18) | 793 |
| 30-45 years | 76(9.2) | 750 (90.79) | 826 |
| 45-60 years | 69(10.52) | 587 (89.48) | 656 |
| 60-75 years | 38(11.99) | 279 (88.01) | 317 |
| >75 years | 10(15.63) | 54 (84.37) | 64 |
| Total | 267(8.89) | 2736(91.10) | 3003 |
While computing the distribution of MRSA in different specialities, Surgery and Orthopaedics departments accounted for most of the cases [Table/Fig-3].
Department wise distribution of MRSA in SSI.
| Departments | MRSA(%) SSI | Non MRSA(%) SSI | Total SSI | |
|---|
| Surgery | 134 (12.49) | 939 (87.51) | 1073 | Chi2 = 51.072 withdegree of freedom5 andp-value <0.001 |
| Orthopedics | 57 (11.85) | 424 (88.15) | 481 |
| Gynaecology | 36 (6.93) | 483 (93.07) | 519 |
| Urology | 15 (3.67) | 347 (96.33) | 362 |
| Pediatrics surgery | 05 (2.25) | 217 (97.74) | 222 |
| Others | 20 (5.7) | 326 (94.23) | 346 |
| 267 | 2736 | 3003 |
*Doxycycline and tigecycline were not used in antibiogram of 73 children [Table/Fig-4].
Antibiotic resistance pattern of Staphylococcus aureus (MRSA and MSSA) isolated from SSI.
| Antibiotics | Strain Total (N) | Resistance n % | Sensitivity n % | Statistics |
|---|
| Clindamycin | MRSA | (267) | 116 | 43.45% | 151 | 56.55% | χ2 8.334df1,p=0.004 |
| MSSA | (782) | 261 | 33.38% | 521 | 66.62% |
| Cefoxitin | MRSA | (267) | 267 | 100.00% | 0 | 0.00 % | Not applicable |
| MSSA | (782) | 1 | 0.13% | 781 | 99.87% |
| Co-trimoxazole | MRSA | (267) | 192 | 71.91% | 75 | 28.09% | χ2 85.826 df 1, p=0.000 |
| MSSA | (782) | 304 | 38.87% | 478 | 61.13% |
| Clarithromycin | MRSA | (267) | 180 | 67.42% | 87 | 32.58% | χ2 78.824 df 1, p=0.000 |
| MSSA | (782) | 275 | 35.17% | 507 | 64.83% |
| Doxycycline* | MRSA | (267) | 74 | 27.72% | 193 | 72.28% | χ2 112.195 df 1,p=0.000 |
| MSSA | (709) | 29 | 4.09% | 680 | 95.95% |
| Fucidin | MRSA | (267) | 20 | 7.49% | 247 | 92.51% | χ2 55.77 df 1, p=0.0001 |
| MSSA | (782) | 0 | 0.00% | 782 | 0.00% |
| Gentamycin | MRSA | (267) | 219 | 82.02% | 48 | 17.98% | χ2 165.726 df 1, p=0.000 |
| MSSA | (782) | 283 | 36.19% | 499 | 63.81% |
| Levofloxacin | MRSA | (267) | 65 | 24.34% | 202 | 75.66% | χ2 18.156 df 1, p=0.000 |
| MSSA | (782) | 102 | 13.04% | 680 | 86.96% |
| Linezolid | MRSA | (267) | 0 | 0.00% | 267 | 100% | Not applicable |
| MSSA | (782) | 0 | 0.00% | 782 | 100% |
| Mupirocin | MRSA | (267) | 31 | 11.61% | 236 | 88.39% | χ2 89.554 df 1, p=0.000 |
| MSSA | (782) | 0 | 0.00% | 782 | 100% |
| Netilmycin | MRSA | (267) | 97 | 36.33% | 170 | 63.67% | χ2 104.830 df 1, p=0.000 |
| MSSA | (782) | 73 | 9.34% | 709 | 90.66% |
| Oxacillin | MRSA | (267) | 246 | 92.13% | 21 | 7.87% | χ2 848.125 df 1, p=0.000 |
| MSSA | (782) | 18 | 2.30% | 764 | 97.70% |
| Tigecycline* | MRSA | (267) | 0 | 0.00% | 267 | 100% | Not Applicable |
| MSSA | (709) | 0 | 0.00% | 709 | 100% |
| Vancomycin.# | MRSA | (267) | 3 | 1.12% | 264 | 98.88% | χ2 5.312 df 1, p=0.021 |
| MSSA | (782) | 0 | 0.00% | 782 | 100% |
* Doxycycline and Tigecycline were not used in antibiogram of children less than 8 years of age. (73 children).
# 17 clinical isolates were resistant to vancomycin 30 μg disc by the disc diffusion test of which 3 strains were confirmed as VISA by E test.
#17 clinical isolates showed resistance to VA (vancomycin – 30 μg disc) by disc diffusion test. Of these, 3 were confirmed as VISA (1.12%) by the E-test showing an MIC range between 4-6 μg ml−1. For the other strains, vancomycin was ≤ 2 μg ml−1indicating Vancomycin sensitive strains. None of the isolates were resistant to Vancomycin by E test (MIC in the range of 16-64 μg ml−1).
The MRSA strains have been found to be 100% sensitive to linezolid and tigecycline in this study. Other highly sensitive drugs were fucidin (92.51%), mupirocin (88.39%), levofloxacin (75.66%) and doxycycline (72.28%) [Table/Fig-4].
The isolated MRSA strains showed high degree of resistance towards clarithromycin, cotrimoxazole and gentamycin in comparison to MSSA strains. Among the 782 cases of MSSA, one was found to be resistant to cefoxitin but sensitive to oxacillin. The Slidex Staph latex agglutination test was also found to be negative with this strain. Therefore, it was considered as Methicillin sensitive Staphylococcus aureus (MSSA).
A decreasing trend of Staphylococcal SSI and MRSA has been found in this study which is statistically significant (χ2 = 16.282, df 3, p =0.001) [Table/Fig-5].
Line chart showing decreasing trend of MRSA over the study period (2009-2012).
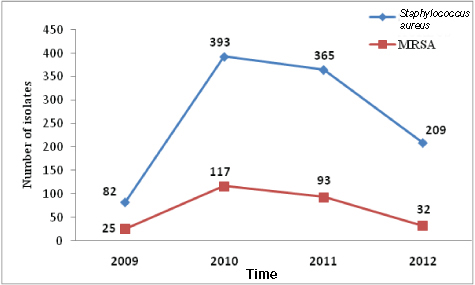
In 2009, rate of MRSA was 30.48% followed by 29.77% in 2010, 25.47% in 2011 and 15.31% in 2012 with an overall MRSA rate of 25.45%. The study revealed presence of mecA gene in 257 strains (96.25 %) and absent in 10 (3.75%) strains.
Discussion
SSI is an important contributor of health care associated infections [25]. It is one of the preventable causes of nosocomial infections [26]. Rate of SSIs in this study was 15.51% which was in tandem with a study in Uttarakhand (17.8%) [27]. But incidence of SSI varied from 2.5-41.9% [28]. Rate of SSI is an important indicator of quality of surgical procedures in a hospital and it is diverse in different set up.
Staphylococcus aureus comprised of more than 1/3rd of SSIs in this study, which was comparable to studies in Gwalior (34%) [29], Karnataka (31.3%) [4] and Uttarakhand (50.4%) [27]. Literature revealed that 80% of healthy individuals across the world harbour Staphylococcus aureus in their skin or anterior nares, and integrity of the skin if breached during any surgery could commonly cause skin and soft tissue infections with this organism [30]. All these factors have made up S.aureus as the most common organism causing SSIs [31]. Among the Gram negative organism causing SSI, E.coli and Klebsiella spp. were the major offenders followed by Pseudomonas spp. and Acinetobacter spp. in this study. E.coli was reported as the most common Gram negative organisms causing SSI in the studies from Uttarakhand [27] and Karnataka [4] also, but Pseudomonas spp. grew up as the second highest Gram negative organism responsible for SSIs in both the studies. Another study revealed Pseudomonas spp. (21%) as the most prevalent organism producing SSIs [29]. So there was a great variation among Gram negative organisms causing SSIs in different geographical areas and set up.
Among the isolated Staphylococcus aureus strains, 25.45% were MRSA which was quite similar to the studies done in Gwalior (27.96%) [29] and Karnataka (28.6%) [4]. Incidence of MRSA in SSI ranged from 15.7 % to 63.5% in studies conducted in India [27] and abroad [32]. A huge variation in incidence of MRSA might depend on pre & postoperative antibiotic policy and surveillance program prevailing in different set up. The incidence of MRSA in the male patients suffering from SSI has been found to be more than the female patients with the male female ratio of 1.67: 1 in this study. There were studies showing male predominance in SSI, though none commented on male: female distribution of MRSA in SSI [4,27].
In the present set up, most of the patients stayed for at least 5-7 days in the hospital during post- operative period in case of major surgeries and wound infection was reported before the patients were discharged from the hospital. Therefore, SSI with MRSA from indoor patients (88.01%) outnumbered the SSI with MRSA cases from outdoor in this study. But with the growing trend of same day surgery and lack of post discharge surveillance, actually contributed to the lesser number of MRSA infected SSIs in outdoor. Since a good number of SSIs might be apparent after discharge from the hospital, a possibility of under reporting could be the reason of paucity in the infected outpatient.
Distribution of SSI infected with MRSA was concentrated (72.28%) among patients aged more than 30 years in this study. Age wise distribution of MRSA cases was proved as statistically significant. Decreasing immunity, low healing power, amplified catabolic processes and existence of co-morbid illnesses, make the older age group more prone to SSIs [33]. But, there was a study reporting the maximum number of SSIs with Staphylococcus, in the age group of 21-40 years [29].
In this study, majority of SSIs with MRSA were from Surgery and Orthopaedics department followed by Obstetrics & Gynaecology which was comparable with a similar type of study in Karnataka [4]. This department wise distribution of MRSA cases was found to be significant.
Overall, MRSA strains were found to be more resistant than MSSA strains to all the antibiotics used and that was statistically significant except for vancomycin, linezolid and tigecycline. Almost similar results were observed in studies in Uttarakhand [27], Gwalior [29] and Karnataka [4] and in a multicentric study from India [34] where vancomycin and linezolid showed 100% sensitivity to MRSA. But no data regarding use of tigecycline was documented in any of these studies. No vancomycin resistant Staphylococcus aureus (VRSA) was detected in this study but 3 (1.12%) isolates were stamped as VISA on the basis of MIC value in E-Test. In a study from Northern India, six (0.76%) VISA strains and two (0.25%) VRSA strains were reported [35]. But in a study from Congo, Africa 19% resistance was seen against vancomycin [32].
The increase in resistance to all group of antibiotics and reports of reduced susceptibility to the glycopeptides made it necessary to search for other alternative drugs for treatment of the MRSA strains [32,35,36]. Antibiogram was done using doxycycline (30μg) and tigecycline (15 μg) discs. They showed encouraging result with 100% sensitivity for tigecycline and 72.28% sensitivity to doxycycline. The sensitivity to tigecycline has been reported to be 100% against MRSA in other studies from India [37,38]. As they were not much used in clinical practice, these two drugs might act as important weapons against MRSA just like vancomycin and linezolid. In this study, newer antibiotics like quinupristin - dalfopristin and daptomycin were not used. Few reports from India had shown encouraging results with these antibiotics [39,40]. As more and more resistance is developing, future work can be directed by performing the in-vitro drug sensitivity testing with these drugs, against MRSA, in this part of India.
Slight fall in the rate of MRSA was noted from 2009 to 2010 (30.48% to 29.77%), followed by a significant fall in the rate during 2011 to 2012 (25.47% to 15.31%) with an overall prevalence of 25.45% during the study period. The fall in the prevalence from 2009-2010 to 2011-2012 was statistically significant (p-value-0.0028). Throughout the world where increasing incidence of MRSA is a threat to the health care system, this study reveals a paradoxical incidence. This has been possible because of effective control measures in the form of proper hand washing, barrier methods of nursing and use of appropriate disinfectants among the health care personnel attending to the patients undergoing surgery. Another important factor for the reduction of prevalence of MRSA is the heightened awareness among doctors and nursing staff about MRSA by thorough campaign, posters and seminars going on in this institute. Fall in prevalence was also observed in a study with strict enforcement of intervention strategy, following an outbreak of MRSA [41]. In our study mecA gene was present in 96.25% strains and absent in 3.75% strains. Similar report of mecA gene negative MRSA has also been reported by Kocazog et al., and Shorman et al., [42,43]. mecA gene negative MRSA was explained by β-lactamase hyperproduction by the isolates [42].
Limitation
1. Sensitivity of newer drugs like quinupristin - dalfupristin and daptomycin, against MRSA have not been demonstrated in this study.
2. Presence of mecB and mecC genes, in mecA negative MRSA strains have not been explored in this current research work.
Conclusion
Staphylococcus aureus played a predominant role in the aetiology of SSIs in this hospital, one fourth of which was due to MRSA strains. Mainstay of treatment for MRSA infections still depends on glycopeptides and linezolid, whereas doxycycline and tigecycline could be used as alternative drugs as revealed in this study. A decline in the rate of MRSA causing SSI in this study might be a ray of hope against the rising trend of this superbug. Compilation of local data on SSIs and feedback are the fundamental concern for the formulation of a proper guideline for peri-operative prophylaxis of antibiotics to mitigate the rate SSIs in the hospital. Post discharge surveillance must now be incorporated in the hospital policy to avoid under reporting of SSIs.
* Doxycycline and Tigecycline were not used in antibiogram of children less than 8 years of age. (73 children).
# 17 clinical isolates were resistant to vancomycin 30 μg disc by the disc diffusion test of which 3 strains were confirmed as VISA by E test.