Introduction
Malaria is a human disease of which causes high morbidity and mortality. In Plasmodium falciparum malaria, the resistance to antimalarial drugs, especially chloroquine (CQ) is one of the paramount factors contributing to the global increase in morbidity and mortality, due to malaria. Hence, there is a need for detection of chloroquine drug resistance genes i.e., pfcrt-o (Plasmodium falciparum chloroquine resistance transporter-o) and pfmdr-1 (Plasmodium falciparum multidrug resistance-1) of P. falciparum and pvcrt-o (Plasmodium vivax chloroquine resistance transporter-o) and pvmdr-1 (Plasmodium vivax multidrug resistance-1) of P. vivax by using molecular methods to prevent mortality in malarial cases.
Aim
To standardize chloroquine drug sensitivity testing by molecular method so as to provide reports of chloroquine within 6-8 hours to physicians for better treatment.
Materials and Methods
This study was conducted over a period of one year from January to December 2014. A Total of 300 blood samples were collected from malaria suspected patient attending MGM Hospital, Kamothe, Navi Mumbai, India. Out of 300 blood samples, 44 were malaria positive as assessed by Thick and Thin blood smear stained, by Leishman’s method and examination with light microscope. Chloroquine drug sensitivity testing was performed using WHO III plate method (micro test). Nested PCR was done for detection of pfcrt-o and pfmdr-1 for P. falciparum and pvcrt-o, pvmdr-1 genes for P. vivax.
Results
Total 44 samples were included in this study, out of which 22 samples confirmed for Plasmodium falciparum and 22 samples confirmed for Plasmodium vivax. Out of 22 P. falciparum 15 (68.18%) samples were chloroquine resistant. P. vivax showed chloroquine resistance to 5 samples (22.73%) by method similar to WHO III plate method (micro test) and nested PCR.
Conclusion
Drug resistance testing by molecular methods is useful for early detection of antimalarial drug resistance. pfmdr-1 along with pfcrt-o can be used as biomarker for chloroquine drug resistance in P. falciparum and pvmdr-1 along with pvcrt-o for P. vivax.
Introduction
The disease Malaria causes high morbidity and mortality in developing countries. According to WHO approximatey 300-500 million malarial cases occur every year, i.e.,90% of the total cases occurring in Africa and Asia. A 700,000 to 2.7 million cases mortality occurr worldwide [1]. According to UNICEF a child dies at every minute from malaria in Africa [2]. A 1.2 billion cases are at risk of malaria, most of whom live in India. However, Southeast Asia contributed 2.5 million cases to the global burden of malaria. Of this, India alone contributed 76% of the total cases [1].
In India around 1.5 million cases of malaria occurr annually. confirmed by clinical examination, radiological examination and laboratory investigations. Chloroquine drug sensitivity can be tested using WHO III plate method [3]. Plasmodium falciparum is responsible for 50% of the total incidence occurring in the world. Chloroquine is the choice of drug for prophylaxis of malaria and maximum cases of malaria by P. falciparum showed resistance to chloroquine which is used as the first line treatment of malaria [4].
Now-a-days the treatment of malaria with chloroquine in P. falciparum cases may cause high morbidity and mortality in patients if treated empirically and without confirmation of the report of antimalarial drug sensitivity testing which is available by both procedures i.e. phenotypic by using WHO III plate (micro test) method and molecular (Polymerase chain reaction) method.
Some studies revealed that chloroquine acts by interfering with heme metabolism in the digestive vacuole of P. falciparum and the drug resistance occurred due to decreased concentration of the drug by efflux pump inhibitor of the parasite [5–7].
Many workers reported that the genetic alterations in P. falciparum are associated with chloroquine drug resistance i.e. P. falciparum multidrug resistance gene (pfmdr-1), and the chloroquine resistance transporter gene pfcrt. Several point mutations in pfmdr-1 gene at positions 754, 1049, 3598, 3622 and 4234 result in amino acid changes at codons 86, 184, 1034, 1042 and 1246, respectively. These amino acid changes have been shown to be associated with chloroquine drug resistance [8–15]. A mutation that occurred in codon 86 (from asparagine to tyrosine, N86Y), involved in the substrate specificity of the gene product (P- glycoprotein), appears to be the most important as this may alter the transport activity of the protein [7]. However, some studies have has reported that the pfmdr-1 gene mutations are also present in chloroquine drug resistance [16].
There are also some variations in point mutations of isolates from different places. The N86Y mutation is present in Southeast Asian chloroquine drug resistant (K1 genotype) isolates whereas it is absent in South American (7G8 genotype) isolates [7].
A study reported that the mutation present in codon 86 has also been evaluated to chloroquine drug resistance in malarial parasites by invitro drug sensitivity testing [16].
The mutations present in the pfcrt-o (codon 74, 75, 76, 220, 271, 326, 371) have also been correlated to chloroquine drug resistance by invitro drug sensitivity testing of P. falciparum in all over the world [17–19]. However, the K76T mutation in pfcrt-o gene has not been observed in chloroquine sensitive strains. It can be regarded as a good molecular bio marker for detection of chloroquine drug resistance in P. falciparum [13,20–22].
In India, especially in the Northeast, the role of mutations in genes pfmdr-1 and pfcrt-o has not been studied in the emergence of P. falciparum chloroquine drug resistance. Studies from other parts of India, reported poor association of chloroquine drug resistance with these gene mutations [23].
In previous days, chloroquine was the recommended first line treatment for uncomplicated malaria in P. falciparum endemic areas. However, now a days this has been changed to artemisinin-based combination therapies. Many malaria-affected areas are still using chloroquine drug for treatment of non-complicated malaria [14,24].
There is a need of chloroquine drug sensitivity testing by molecular methods for better treatment of malaria. Malarial parasite population control, genetic studies and determination of the presence of chloroquine drug sensitivity is needed to control the burden of the disease [25].
Materials and Methods
This prospective and analytical study was conducted at Department of Microbiology and Central Research Laboratory, MGM Medical College and Hospital over a period of one year from January to December 2014. Total 300 blood samples were collected from malaria suspected patients attending MGM Hospital with symptoms of fever and chills. Patients already on antimalarial treatment were excluded from the study. Out of 300 blood samples 44 were malaria positive. 22 P. falciparum and 22 P. vivax.
For drug sensitivity and molecular analysis, approximately 5 ml of venous blood was collected from the malaria suspected patients (1 ml for thick and thin smear, 2 ml for invitro antimalarial drug sensitivity testing by WHO III plate method and 2 ml for DNA extraction for detection of drug resistance genes by Nested PCR) who were tested positive for Plasmodium falciparum using thick and thin blood smear and stained with Leishman’s stain. The blood was stored in cryo vials and stored in at –20°C. The study protocol was reviewed and approved by the Ethical Review Committee of MGM Institute of Health Sciences (Deemed University), Navi Mumbai. Informed written consent was obtained from the patients before start the study.
In vitro drug sensitivity testing: Antimalarial drug sensitivity testing was performed by invitro micro test (Mark III) according to Singh et al., [1] chloroquine drug sensitivity test was performed immediately after the collection of blood. The test was considered valid and interpretable if 10% of the parasites in the control well (drug free well) had developed into the schizonts after 24–36 hours incubation. Isolates were considered resistant if they showed schizont maturation at chloroquine concentrations 8 pmol/well (1.6 mmol/L blood). To evaluate the drug-parasite response, the EC50 value (50% inhibition) was calculated by HN Non Lin (V. 1.01 Beta) Software [26].
DNA extraction: The DNA extraction form above samples was performed by using DNA Mini Kit (Invitrogen) spin column method.
Primer Design: Primers used in this study were designed from published articles and were procured from Eurofins Genomics India [Table/Fig-1].
Primers for nested PCR for detection of drug resistant gene in malaria parasites.
| Sr. | Name oforganism | Gene | Sequences 5’ – 3’ | Gene code | Ref. |
|---|
| 1 | P. vivax | pvmdr1 | F-GCGAACTCGAATAAGTACTCCCTCTA | EU333979.1 | [27] |
| R-GGCGTAGCTTCCCGTAAATAAA | EU333979.1 |
| pvcrt-o | F-CGCTGTCGAAGAGCC | EU333972.1 | [28] |
| R- AGTTTCCCTCTACACCCG | EU333972.1 |
| 2 | P. falciparum | pfmdr1 | F- TGTATGTGCTGTATTATCAGGAGGAAC-3′ | JN578609.1 | [29] |
| R-AATTGTACTAAACCTATAGATACTAATGATAATATTATAGG | JN578609.1 |
| pfcrt-o | F - TGAGAATTAGATAATTTAGTACAAGAAGGAA | JF520758.1 | [17] |
| R- CGTGAGCCATCTGTTAAGGTC | AF030694.2 |
Optimization of DNA preparation: DNA was extracted from 200 μl of blood in EDTA using the DNA extraction kit (Invitrogen, USA) spin column method and stored at 4°C until PCR could be completed. DNA used for the PCR was standardized through DNA Mini Kit (Invitrogen).
Polymerase Chain Reaction
Nested PCR amplifications were performed in accordance to the procedure as followed by Stephanie P. Johnston et al., within the cycling parameters [Table/Fig-2] by using a PeqSTAR 96Xx Universal Gradient PCR thermal cycler (Peqlab, Germany). According to the procedure master mix “BioMix Red” (Bioline, India), 5μl DNA, 10 pmol of primers were added and mixed to obtain 50μl final volume of the PCR mix. The PCR products along with the appropriate ladder (Bioline, India) and known positive and negative samples from previous malaria diagnosed or uninfected individuals used as controls were subjected to electrophoresis in a 1.5% agarose gel using 1X Tris Acetate EDTA (TAE) buffer. The gel was then placed on the surface of the UV transilluminator (BioEra, India) and visualized in dark. The DNA bands were documented by gel documentation system (BioEra, India).
Cycling conditions of PCR reactions for detection of drug resistance gene of P. falciparum and P. vivax.
| Reactiona | Cycling conditions |
|---|
| pvmdr-1 | Initial denaturation at 94°C for 10 minutes followed by 35 cycles of denaturation at 94°C for 50 seconds, annealing at 62°C for 1 min, and extension at 72°C for 1 min 30 seconds [30]. |
| pvcrt-o | Initial denaturation at 94°C for 10 minutes followed by 35 cycles of denaturation at 94°C for 50 seconds, annealing at 61°C for 1 min, and extension at 72°C for 1 min 30 seconds [30]. |
| pfmdr-1 | Initial denaturation at 50°C for 2 minutes, 95°C for 10 minutes followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 1 minutes and extension at 72°C for 1 minute [31]. |
| pfcrt-o | Initial denaturation at 94°C for 10 minutes followed by 40 cycles of denaturation at 94°C for 1 minute, annealing at 56°C for 1 min, and extension at 72°C for 1 min 30 seconds [32]. |
Results
Total 44 malaria positive samples were included in the study, out of which 22 were P. falciparum and 22 were P. vivax.
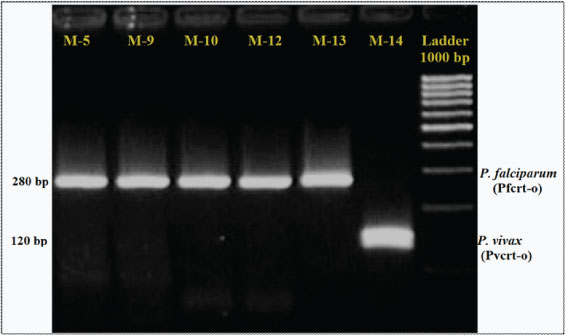
A 15 out of 22 Plasmodium falciparum multiplied and grew rapidly in RPMI-1640 medium supplemented with O positive red blood cells under chloroquine pre-coated microtitre plate which were regarded as resistant to chloroquine and 7 out of 22 where sensitive to chloroquine drug. Electrophoresis gel picture shows band of pfcrt-o 280 bp and pvcrt-o 120bp [Table/Fig-3].
Gel picture showing band of pfcrt-o (Plasmodium falciparum chloroquine resistant transporter-o) 280 bp and pvcrt-o (Plasmodium vivax chloroquine resistant transporter-o) 120 bp.
A 5 out of 22 Plasmodium vivax multiplied and grew in McCoy 5A medium supplemented with reticulocytes under chloroquine pre-coated microtitre plate which were regarded as resistant to chloroquine and 17 out of 22 where sensitive to chloroquine drug [Table/Fig-4,5].
Detection of drug resistance genes of P. falciparum by PCR.
| No. of samples tested | pfmdr-1 | pfcrt-o |
|---|
| Control strain CQS* 3D7 (n=1) | 0 | 0 |
| Patient samples (n=22) | 15 | 15 |
Detection of drug resistance genes of P. vivax by PCR. CQS* chloroquine sensitive.
| No. of samples tested | pfmdr-1 | pfcrt-o |
|---|
| Positive Control strain CQS* (n=1) | 0 | 0 |
| Patient samples (n=22) | 05 | 05 |
PCR amplification of extracted DNA of P. falciparum and P. vivax revealed that chloroquine drug resistant genes (pfcrt-o and pfmdr-1) were detected in 15 out of 22 P. falciparum. However, 5 out of 22 chloroquine drug resistant genes (pvcrt-o and pvmdr-1) were detected in P. vivax.
Discussion
Increasing drug-resistance in malarial parasites especially in Plasmodium falciparum to chloroquine has created major health problem in the world [18]. Now-a-days in malaria endemic areas chloroquine is taken as prophylactic drug only but still it showed good response in case of vivax malaria. However, the drug sensitivity by moleculars method could help to provide better treatment to patients [1].
The fast rate of emergence of chloroquine drug resistance has become a major burden during malaria control. chloroquine resistance in P. vivax was noted for the first time in Papua New Guinea [33] and from there it has spread to other parts of the world. From India also there are now several reports of chloroquine resistance in P. vivax [34–36]. Resistance in P. vivax is more serious as hypnozoites will cause relapse of resistant parasites and P. vivax is a mixture of various strains with respect to incubation period, relapsing pattern and response to primaquine [37] since sulpha drugs are not effective in its treatment.
The development of molecular methods for detection of drug resistant genes in malarial parasites has very important role for screening of the drug resistance, and providing better treatment to patients.
We describe a Nested PCR assay to detect drug resistance genes of P. falciparum (pfcrt-o and pfmdr-1) and P. vivax (pvcrt-o and pvmdr-1). We could successfully find chloroquine drug resistant genes in 15 of the 22 P. falciparum and 7 of the 22 P. vivax in Navi Mumbai, India. This area has big mountains and becomes malaria endemic during rainy season because it favours mosquito breeding.
In this study, all 44 samples were subjected to PCR for amplification of pfmdr-1 and pfcrt-o for P. falciparum and pvmdr-1 and pvcrt-o for P. vivax. pfmdr-1 and pvmdr-1 are multidrug resistance genes for P. falciparum and P. vivax. pfcrt-o and pvcrt-o are chloroquine drug resistance genes for P. falciparum and P. vivax.
pfmdr-1 and pfcrt-o genes were not detected in 3D7 control strain which is P. falciparum chloroquine sensitive strain. However, 15/22 (68.18%) patient samples showed presence of both pfcrt-o and pfmdr-1 genes confirming chloroquine drug resistance by molecular methods. In vito analysis of PCR and chloroquine susceptibility of pfmdr-1 and pfcrt-o polymorphisms in P.falciparum have revealed that chloroquine resistance has been linked to the mutations in pfcrt and pfmdr1 genes [38].
pvmdr-1 and pvcrt-o mutant genes were not detected in chloroquine sensitive strain of P. vivax by in vitro drug sensitivity testing. However, 5/22 (22.73%) patient samples showed presence of both pvcrt-o and pvmdr-1 genes confirming chloroquine drug resistance. This validates the findings of invitro drug sensitivity testing. Findings from a previous study suggests that increased expression levels of the pvcrt o and pvmdr-1 genes are strongly associated with clinical severity and chloroquinone resistance in P.vivax infections [39]. pvmdr1 Y976F mutation has been identified as a possible genetic marker for chloroquine resistance in P.vivax. in a study that has investigated the association between the polymorphisms of pvmdr1 and pvcrt o as markers of chloroquine resistance. pvmdr 1 Y976F mutation was detected only in 7/30(23.3%) P.vivax isolates. Chloroquine resistance phenotype in P.falciparum are strongly associated with the point mutations in pfcrt genes especially K76T [40].
In our study, 68.18% samples showed presence of pfcrt-o drug resistance genes. This finding is closer to Shrivastava SK et al., from India (86.95%), Sutar SKD et al., from Odisha, India (80%) and, Anvikar AR et al., from India (90.47%) [32,41,42]. Lim P et al., from Cambodia reported in all isolates of chloroquine resistance [43]. Babiker HA et al., from Sudan reported association of chloroquine resistance transporter gene (pfcrt-o) with in vivo and in vitro resistance [44]. Jalousian F et al., from Iran however reported less value of 23.1% [45]. Chloroquine in combination with primaquine or alone is still effective against P.vivax malaria according to a study from Kolkata in which the in vivo efficacy of chloroquine (CQ) and chloroquine plus Primaquine was determined [46]. For comparison of genetic determinants of chloroquine resistance in both P.falciparum and P.vivax, identification of difference in the orthologous genes is important. The development of resistance may be different in both P.falciparum and P.vivax however the mechanism of chloroquine resistance is probably similar in both of these two species [40].
In our study, we found 22.73% resistant genes of pvcrt-o and pvmdr-1. However, Lu F et al., from Central China, they did not find pvcrt-o and pvmdr-1 resistant genes [30]. A direct relationship between unusual mutation in pfmdr-1 and pfcrt genes and prevalence of chloroquine resistance has been determined with early treatment failure cases [47] chloroquine resistance monitoring through molecular markers is thus a useful tool that can be used for future control [40]. Such molecular markers that are associated with the increase in chloroquine resistance and disease severity in P.vivax are needed [39].
Limitations
In this study we could not study resistance in other drugs like Artemisinin, Mefloquine, Quinine, Sulphadoxine/Pyrimethamine and Primaquine.
Thus, further studies are required to enable the detection of resistance against these drugs, which can help in the treatment of multi drug resistance cases of malaria. Nevertheless, given the advantages of this method, nested PCR could serve as a useful tool for detection chloroquine drug resistant in malaria in endemic area.
Conclusion
Drug sensitivity testing by molecular methods is useful for early detection of drug resistance of chloroquine and will help physician to provide better treatment which decreases morbidity and mortality of patients. pfmdr-1 along with pfcrt-o can be used as biomarker for chloroquine drug resistance in P. falciparum and pvmdr-1 along with pvcrt-o for P. vivax.
[1]. Kumar A, Valecha N, Jain T, Dash AP, Burden of Malaria in India: Retrospective and Prospective ViewAm J Trop Med Hyg 2007 77(6):69-78. [Google Scholar]
[2]. http://www.unicef.org/prescriber/eng_p18.pdf [Google Scholar]
[3]. Singh G, Urhekar AD, Raksha Invitro Antimalarial Drug Sensitivity Testing For Plasmodium falciparum and Plasmodium vivaxIOSR Journal of Dental and Medical Sciences 2015 14(4):49-55. [Google Scholar]
[4]. National Drug Policy on Malaria (2013) Directorate General of National Vector Borne Disease Control Programme, Ministry of Health & Family Welfare, Government of India. New Delhi: 1–15 [Google Scholar]
[5]. Fitch CD, Plasmodium falciparum in owl monkeys: drug resistance and chloroquine binding capacityScience 1970 169(942):289-90. [Google Scholar]
[6]. Douki JBL, Boutamba SDD, Zatra R, Edou SEZ, Ekomy H, Increased prevalence of the Plasmodium falciparumpfmdr1 86N genotype among field isolates from France ville, Gabon after replacement of chloroquine by artemether–lumefantrine and artesunate–mefloquineInfect Gen Evo 2011 11:512-17. [Google Scholar]
[7]. Sanchez CP, Dave A, Stein WD, Lanzer M, Transporters as mediators of drug resistance in Plasmodium falciparumInt J Parasitol 2010 40:1109-18. [Google Scholar]
[8]. Foote SJ, Kyle DE, Martin RK, Oduola AM, Forsyth K, Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparumNature 1990 345(6272):255-58. [Google Scholar]
[9]. Basco LK, Le Bras J, Rhoades Z, Wilson CM, Analysis of pfmdr1 and drug susceptibility in fresh isolates of Plasmodium falciparum from sub-Saharan AfricaMol Biochem Parasitol 1995 74(2):157-66. [Google Scholar]
[10]. Cox-Singh J, Singh B, Alias A, Abdullah MS, Assessment of the association between three pfmdr1 point mutations and chloroquine resistance invitro of Malaysian Plasmodium falciparum isolatesTrans R Soc Trop Med Hyg 1995 89(4):436-37. [Google Scholar]
[11]. Adagu IS, Dias F, Pinheiro L, Rombo L, do Rosario V, Warhurst DC, Guinea Bissau: association of chloroquine resistance of Plasmodium falciparum with the Tyr86 allele of the multiple drug-resistance gene pfmdr1Trans R Soc Trop Med Hyg 1996 90(1):90-91. [Google Scholar]
[12]. Duraisingh MT, Drakeley CJ, Muller O, Bailey R, Snounou G, Evidence for selection for the tyrosine-86 allele of the pfmdr 1 gene of Plasmodium falciparum by chloroquine and amodiaquineParasitol 1997 114:205-11. [Google Scholar]
[13]. Chaijaroenkul W, Ward SA, Mungthin M, Sequence and gene expression of chloroquine resistance transporter (pfcrt) in the association of invitro drugs resistance of Plasmodium falciparumMalaria J 2011 10:42 [Google Scholar]
[14]. Murambiwa P, Masola B, Govender T, Anti-malarial drug formulations and novel delivery systems: A reviewActa Tropica1 2011 18:71-79. [Google Scholar]
[15]. Atroosh WM, Mekhlafi HM, Mahdy MAK, Surin J, The detection of pfcrt and pfmdr1point mutations as molecular markers of chloroquine drug resistance, Pahang, MalaysiaMalaria J 2012 11:251 [Google Scholar]
[16]. Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1Antimicrob Agents Chemother 2003 47:2418-23. [Google Scholar]
[17]. Fidock DA, Nomura T, Talley AK, Mutations in the P. falciparum digestive vacuole transmembrane protein pfcrt and evidence for their role in chloroquine resistanceMol Cell 2000 6(4):861-71. [Google Scholar]
[18]. Sidhu AB, Verdier-Pinard D, Fidock DA, chloroquine resistance in Plasmodium falciparum malaria parasites conferred by pfcrt mutationsScience 2002 298(5591):210-13. [Google Scholar]
[19]. Vathsala PG, Pramanik A, Dhanasekaran S, Widespread occurrence of the Plasmodium falciparum chloroquine resistance transporter (pfcrt) gene haplotype SVMNT in P. falciparum malaria in IndiaAm J Trop Med Hyg 2004 70(3):256-59. [Google Scholar]
[20]. Ojurongbe O, Ogungbamigbe TO, Beyioku AFF, Rapid detection of pfcrt and pfmdr1 mutations in Plasmodium falciparum isolates by FRET and in vivo response to chloroquine among children from Osogbo, NigeriaMalaria J 2007 6:41 [Google Scholar]
[21]. Mekhlafi AMA, Mahdy MAK, Mekhlafi HM, High frequency of Plasmodium falciparum chloroquine resistance marker (pfcrt T76 mutation) in Yemen: An urgent need to re-examine malaria drug policyParasit Vect 2011 4:94 [Google Scholar]
[22]. Veiga MI, Ferreira PE, Jornhagen L, Novel polymorphisms in Plasmodium falciparum ABC transporter genes are associated with major ACT antimalarial drug resistancePLoS ONE 2011 6(5):e20212 [Google Scholar]
[23]. Bhattacharya PR, Pillai CR, Strong association, but incomplete correlation, between chloroquine resistance and allelic variation in the pfmdr-1 gene of Plasmodium falciparum isolates from IndiaAnn Trop Med Parasitol 1999 93(7):679-84. [Google Scholar]
[24]. Ranjitkar S, Schousboe ML, Thomsen TT, Prevalence of molecular markers of anti-malarial drug resistance in Plasmodium vivax and Plasmodium falciparum in two districts of NepalMalaria J 2011 10:75 [Google Scholar]
[25]. Alam MT, Souza DK, Vinayak S, Selective sweeps and genetic lineages of Plasmodium falciparum drug -resistant alleles in GhanaJ Infect Dis 2011 203:220-27. [Google Scholar]
[26]. http://www.meduniwien.ac.at/user/harald.noedl/malaria/download.html [Google Scholar]
[27]. Lekweiry LM, Boukhary AOMS, Gaillard T, Wurtz N, Bogreau H, Hafid JE, Molecular surveillance of drug-resistant Plasmodium vivax using pvdhfr, pvdhps and pvmdr1 markers in Nouakchott, MauritaniaJ Antimicrob Chemother 2011 :1-8.doi:10.1093/jac/dkr464 [Google Scholar]
[28]. Suwanarusk R, Russell B, Chavchich M, Chalfein F, Kenangalem E, chloroquine Resistant Plasmodium vivax: Invitro Characterisation and Association with Molecular PolymorphismsPLoS ONE 2007 2(10):e1089doi:10.1371/journal.pone.0001089 [Google Scholar]
[29]. Purfield A, Nelson A, Laoboonchai A, Congpuong K, McDaniel P, Miller RS, New method for detection of pfmdr1 mutations in Plasmodium falciparum DNA using real-time PCRMalaria J 2004 3(9) [Google Scholar]
[30]. Lu F, Wang B, Cao J, Sattabongkot J, Zhou H, Zhu G, Prevalence of drug resistance-associated gene mutations in Plasmodium vivax in Central ChinaKorean J Parasitol 2012 50(4):379-84. [Google Scholar]
[31]. Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, Resistance to antimalarials in Southeast Asia and Genetic polymorphisms in pfmdr 1Antimicrob agents Chemother 2003 47(8):2418-23. [Google Scholar]
[32]. Shrivastava SK, Gupta RK, Mahanta J, Dubey ML, Correlation of Molecular Markers, pfmdr1-N86Y and pfcrt-K76T, with Invitro chloroquine Resistant Plasmodium falciparum, Isolated in the Malaria Endemic States of Assam and Arunachal Pradesh, Northeast IndiaPLoS ONE 2014 9(8):e103848doi:10.1371/journal.pone.0103848 [Google Scholar]
[33]. Rieckman KH, Davis DR, Hutton DC, Plasmodium vivax resistance chloroquine?Lancet 1989 2:1183-84. [Google Scholar]
[34]. Potkar CN, Kashersagar NA, Kathwria R, Resurgence of malaria and drug resistance in P. falciparum and P. vivax species in BombayJ Assoc Phys India 1994 43:336-38. [Google Scholar]
[35]. Garg N, Gopinath P, Bodhe P, Kshersagar A, Vivax malaria resistant to chloroquine: case reports from BombayTrans R Soc Trop Med Hyg 1995 89:656-57. [Google Scholar]
[36]. Dua VK, Kar PK, Sharma VP, Chloroquine resistant P. vivax in IndiaTrop Med Intl Hlth 1996 1:816-19. [Google Scholar]
[37]. Adak T, Sharma VP, Orlov VS, Studies on P. vivax relapse pattern in DelhiAm J Trop Med Hyg 1978 59:175-79. [Google Scholar]
[38]. Thomas SM, Ndir O, Invitro chloroquine susceptibility and PCR analysis of pfcrt and pfmdr1 polymorphisms in Plasmodium falciparum isolates from SenegalAm J Trop Med Hyq 2002 66(5):474-80.(http://www.ncbi.nlm.nih.gov/pubmed/12201579) [Google Scholar]
[39]. Melo GC, Monteiro WM, et al. Expression levels of pvcrt-o and pvmdr-1 are associated with chloroquinone resistance and severe P.vivax malaria in patients of the Brazilian Amazon – (http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0105922) [Google Scholar]
[40]. Rungsihirunrat K, Muhamad P, Plasmodium vivax drug resistance; pvmdr1 and pvcrt-o polymorphisms in relation to chloroquine sensitivity from a malaria endemic area of ThailandKorean J Parasitol 2015 53(1):43-9.(http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4384798/) [Google Scholar]
[41]. Sutar SKD, Gupta B, Ranjit M, Kar SK, Das A, Sequence analysis of coding DNA fragments of pfcrt and pfmdr-1 genes in Plasmodium falciparum isolates from Odisha, IndiaMem Inst Oswaldo Cruz, Rio de Janeiro 2011 106(1):78-84. [Google Scholar]
[42]. Anvikar AR, Sharma B, Sharma SK, Ghosh SK, Bhatt RM, Invitro assessment of drug resistance in Plasmodium falciparum in five States of IndiaIndian J Med Res 2012 135:494-99. [Google Scholar]
[43]. Lim P, Chy S, Ariey F, Incardona S, Chim P, Sem R, pfcrt Polymorphism and chloroquine Resistance in Plasmodium falciparum Strains Isolated in CambodiaAntimicrob. Agents Chemother 2003 47(1):87-94. [Google Scholar]
[44]. Babiker HA, Pringle SJ, Abdel-Muhsin Mackinnon AM, Hunt P, Walliker D, High-Level chloroquine Resistance in Sudanese Isolates of Plasmodium falciparum Is Associated with Mutations in the chloroquine Resistance Transporter Gene pfcrt and the Multidrug Resistance Gene pfmdr1The Journal of Infectious Diseases 2001 183:1535-38. [Google Scholar]
[45]. Jalousian F, Dalimi A, Samiee SM, Ghaffarifar F, Soleymanloo F, Naghizadeh R, Mutation in pfmdr1 gene in chloroquine-resistant Plasmodium falciparum isolates, Southeast IranInternational Journal of Infectious Diseases 2008 12:630-34. [Google Scholar]
[46]. Guanguly S, Saha P, et al. In vivo therapeutic efficacy of chloroquine alone or in combination with primaquine against viavx malaria in pvmdr1 and pvcrt o genes. (http://www.ncbi.nlm.nih.gov/pubmed/23262997) [Google Scholar]
[47]. Das S, Association between prevalence of chloroquine resistance and unusual mutations in pfmdr-1 and pfcrt genes in IndiaAm J Trop Med Hyg 2013 88(5):828-34.(https://www.researchgate.net/publication/236061526_Association_between_Prevalence_of_chloroquine_Resistance_and_Unusual_Mutation_in_pfmdr-I_and_pfcrt_Genes_in_India) [Google Scholar]