Typhoid fever is a major cause of morbidity and mortality with an estimated global incidence of 21.6 million cases and 216, 510 death per year, it is endemic in many developing countries particularly the Indian subcontinent, South and Central America [1].
Reduced susceptibility to ceftriaxone and ciprofloxacin has been reported previously for S. Typhi and S. Paratyphi A in Kuwait [7]. Many reports suggest that ciprofloxacin usage should be re-considered in treating Salmonella infections due to reduced susceptibility. However there is a decline in MDR strains of S. Typhi and S. Paratyphi A [8].
Resistance to quinolones commonly arises via mutations in the genes encoding DNA gyrase (gyrA and gyrB) and DNA topoisomerase IV (parC and parE) [9]. However, in Salmonella species, mutations associated with quinolone resistance are mostly present in the QRDR of the gyrA gene [10,11]. The most commonly reported mutation is at Ser 83, which also has a site for HinfI restriction [10].
Nalidixic acid susceptibility can be used as a possible marker for the detection of reduced susceptibility to ciprofloxacin and ciprofloxacin MIC in Nalidixic Acid-Resistant Strains (NARST) was about 10-fold higher than that of Nalidixic Acid-Sensitive Strains (NASST) [12]. To avoid the delay in appropriate therapy and decrease the associated morbidity and mortality of enteric fever, there is a need to identify the Salmonella strains likely to cause an infection not responding to ciprofloxacin therapy.
Materials and Methods
Collection of Clinical Salmonella Isolates
A total of 133 S. Typhi isolates (131 were blood isolates, one from stool and one from bone marrow aspirate) were isolated and identified by standard microbiological techniques during 2007 – 2009 (first part of the study) and confirmed by slide agglutination test using commercially available anti-sera (KIPM, Chennai) and stored (Glycerol broth at -20 degree Celsius) for further use.
Minimum Inhibitory Concentration (MIC) test for ampicillin, nalidixic acid, ciprofloxacin, cefotaxmie and ceftriaxone were performed by agar dilution method according to CLSI 2014 guidelines using commercially available antibiotics (Himedia, Mumbai) [13]. A part of this study was already published [14] whereas in the present study which has been conducted from October 2011 to December 2014 we have selected the Salmonella isolates based on the MIC for quinolone and fluoroquinolone antibiotics and analysed for mutation in QRDRs.
Preparation of Template DNA: A single bacterial colony from overnight culture was suspended in 100 μL of 50 mM concentration of NaOH and kept in a water bath at 97°C for 3 minutes and then placed in to the refrigerator at 4°C for 5 minutes. After refrigeration 16 μL of 1M of Tris-HCl was added and centrifuged at 8000 rpm for 2 minutes. The supernatant was transferred and stored at -20°C [15].
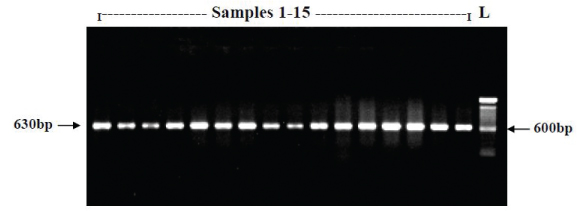
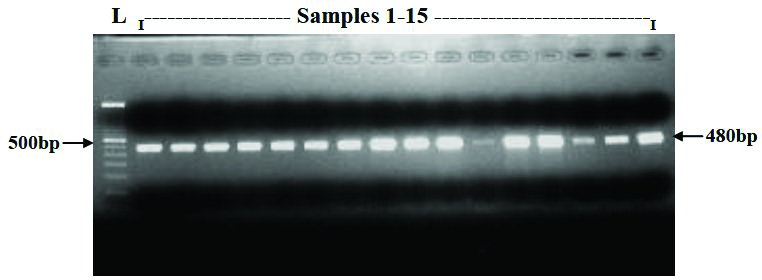
PCR Amplification of gyrA and parC genes: Template DNA prepared from bacterial strains as described above was amplified by PCR using oligonucleotides procured from Sigma-USA. The following primers were adapted from Renuka et al., 2004, GyrA F: 5’ ATG AGC GAC CTT GCG AGA GAA ATT ACA CCG 3’ and GyrA R: 5’ TTC CAT CAG CCC TTC AAT GCT GAT GAT GTC TTC 3’ for gyrA (630bp). The parC gene sequence was retrieved from NCBI database and sequence specific primers were designed using Primer3 online tool ParC F: 5’ATG AGC GAT ATG GCA GAG CGC CTT GCG CTA 3’ and ParC R: 5’ ACG CGC CGG TAA CAT TTT CGG TTC CTG CAT 3’ for parC (480bp). The primers were procured from Sigma Oligos, India. PCR was performed by using Gene Amp 9700 thermal cycler (ABI USA) in final reaction volume of 50 μL containing 5 μL of 10x PCR buffer with MgCl2 {NEB, USA}, 5 μL of 10mM dNTP Mix (Takara, Japan), 2 μL of 2μM forward and reverse primers, 0.25 μL of Taq DNA polymerase (5U/μL) {NEB, USA}, 2.5 μL of template DNA and 35.25 μL of PCR water.
The thermal conditions for the amplification of both gyrA and parC gene comprised one cycle of 2 min at 95°C, followed by 35 cycles of 45 sec at 94°C, 45 sec at 60°C and 30 sec at 72°C, with a final extension of 7 min at 72°C. The 10 μL of PCR product was resolved in 1% (w/v) agarose gel (Sigma, India) prepared in 0.5X Tris Acetic acid EDTA (TAE) buffer and detected by ethidium bromide staining after electrophoresis (BioRad, USA). The 1000bp DNA ladder (Gibco BRL, USA) was used to refer the size of the PCR product in the gel. The gel was documented using gel documentation system (BioRad, USA).
HinfI Restriction Digestion for gyrA gene: The amplified 630bp fragment of the gyrA gene has three HinfI restriction sites one of which lies at Ser83. The PCR product was digested for the detection of gyrA Ser83 mutation by HinfI enzyme. A volume of 20 μL of precipitated PCR product was digested with 10 units of HinfI (New England Biolab-USA) at 37 °C for 16hrs.
The tubes were short spun after the complete digestion. Two percent agarose gel with ethidium bromide (50μg/ml) was prepared using 0.5xTAE buffer and 10 μL of the HinfI digested PCR product was mixed with 2 μL of 6x gel loading dye and resolved at 100V for 25 minutes using 0.5xTAE as a tank buffer; 5 μL of 1000bp DNA ladder (Gibco Brl-USA) was used to refer the size of the digested fragments in the gel. Using gel documentation system (Bio Rad-USA), the gel was documented.
TSP5091 Restriction Digestion for parC: The 480bp PCR product was subjected to restriction digestion using TSP5091enzyme, it has three restriction sites, one of which lies at Trp106-Gly. The PCR product was performed for the detection of parC Trp106 mutation by TSP5091 restriction digestion. A volume of 10μL of precipitated parC PCR product was digested by 10 units of TSP5091 (New England Biolab-USA) at 65°C for 16 hours.
After the complete digestion the tubes were short spun. Two percent agarose gel with ethidium bromide (50μg/ml) prepared using 0.5xTAE buffer and 10 μL of the TSP5091 digested PCR product was mixed with 2 μL of 6x gel loading dye and resolved at 100V for 25 minutes using 0.5xTAE as a tank buffer; 5 μL of 1000bp DNA ladder (Gibco Brl-USA) was used to refer the size of the digested fragments in the gel. The gel was documented using gel documentation system (Bio Rad-USA).
DNA Sequencing for gyrA and parC: Four Salmonella isolates selected based on the MIC of ciprofloxacin (0.06μg/ml to 0.5μg/ml) and nalidixic acid (16μg/ml to 256μg/ml). The gyrA and parC genes of the representative strains were amplified by PCR and nucleotide sequencing was performed (Macrogen inc. Seoul, Korea).
Results
A total of 133 S. Typhi (131 were blood isolates, one from stool and one from bone marrow aspirate) were included in this study. MIC test has been widely considered as a standard method in determining the antibiotic susceptibility of an organism. Hence, we used the MIC test results [Table/Fig-1] to analyse various parameters in this study.
MICs of antibiotics for the S. Typhi isolates.
| Drugconcentration(μg/ml) | Nalidixicacid | Ciprofloxacin | Ampicillin | Cefotaxime | Ceftriaxone |
|---|
| 256 | 104 | - | - | - | - |
| 128 | 14 | - | - | - | - |
| 64 | 7 | - | - | - | - |
| 32 | 7 | - | 4 | - | - |
| 16 | - | - | 1 | - | - |
| 8 | - | - | 1 | - | - |
| 4 | 1 | - | 25 | - | - |
| 2 | - | - | 95 | - | - |
| 1 | - | 28 | 7 | - | - |
| 0.5 | - | 25 | - | 36 | 32 |
| 0.25 | - | 58 | - | 79 | 62 |
| 0.12 | - | 16 | - | 12 | 30 |
| 0.06 | - | 6 | - | 6 | 9 |
| 0.03 | - | - | - | - | - |
-, No growth
The CLSI (2014) interpretive criteria for sensitive, intermediate and resistant strains, respectively, are (mg/ml): nalidixic acid, ≤16, -, ≥32; ciprofloxacin, ≤0.06, 0.12-0.5, ≥1; ampicillin, ≤8, 16, ≥32; cefotaxime, ≤1, 2, ≥4; ceftriaxone, ≤1, 2, ≥4.
In our study only one out of 133 isolates (0.8%) of S. Typhi was found to be nalidixic acid sensitive and 99.2% (132/133) were resistant by MIC studies. Ciprofloxacin resistance was observed in 21% (28/133) of isolates. About 96.2% (128/133) isolates were sensitive to ampicillin by MIC studies; 1/133 was intermediate and only 4/133 was found to be resistant (32 μg/ml). 100% sensitivity was observed for cefotaxime and ceftriaxone.
PCR –RFLP for gyrA and parC nucleotide sequence analysis: HinfI restriction digestion analysis of gyrA PCR Product
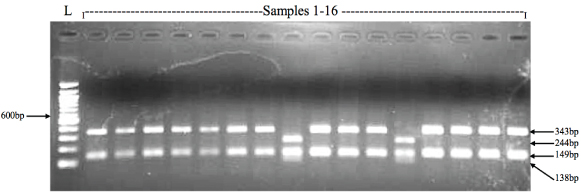
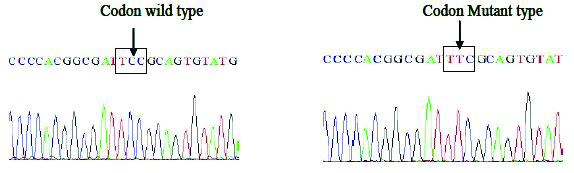
The 630bp PCR fragment of gyrA gene has three HinfI restriction sites, one of which present at the codon 83 serine (TCC), when mutated to phenylalanine (TTC) it leads to the loss of the restriction site. HinfI restriction digestion analysis of the gyrA PCR product showed that 94% (125/133) of S. Typhi strains in the present study were positive for Ser83 mutation, whereas only 6% (8/133) were wild type (Ser83). HinfI cuts the 630bp of gyrA gene product at three sites therefore the restriction pattern of gyrA gene product from nalidixic acid resistant strains with HinfI digestion yield three fragments of sizes 343bp, 149bp and 138bp [Table/Fig-2,3].
Representative document of agarose gel representing the gyrA PCR product (630bp).
Representative RFLP electrophoretic gel showing the gyrA PCR product digested with HinfI.
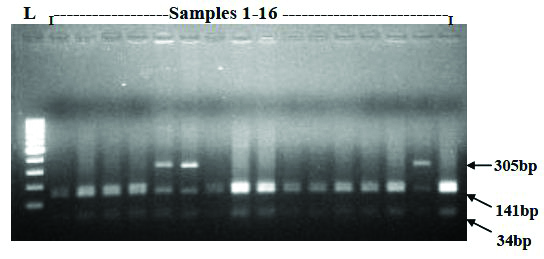
TSP509I restriction digestion analysis of parC PCR Products: The 480bp PCR product of parC gene has three Tsp509I restriction sites, one of which is removed by Trp106-Gly mutation (TGG-GGG) in S. Typhi. The parC PCR product digested with Tsp509I revealed that only 21.8% (29/133) were harbouring the Trp106-Gly mutation; while 78.2% (104/133) were wild type Trp106 [Table/Fig-4,5].
Representative document of agarose gel showing the parC PCR product (480bp).
Representative RFLP electrophoretic gel showing the parC PCR product digested with Tsp5091.
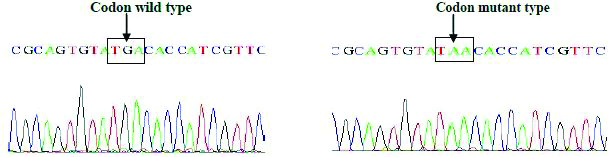
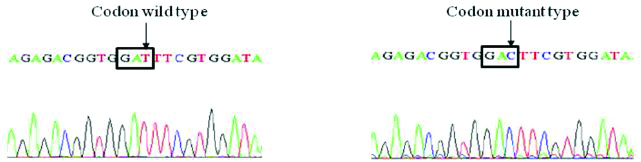
Sequencing BLAST results of gyrA and parC genes of Salmonella isolates: Four S. Typhi isolates were selected and their respective gyrA and parC genes were amplified by PCR and sequenced by using sequencing service (Macrogen Inc. Seoul, Korea). The Basic Local Alignment Search Tool (BLAST) analysis of the sequence results confirmed the gyrA Ser83Phe mutation as observed in RFLP experiment, another non-synonymous mutation GAC-AAC at codon 87 of gyrA (Asp87Asn) was observed in the DNA sequencing results of two S. Typhi strains [Table/Fig-6a,b and c].
Sequence and BLAST result of GyrA mutation at Ser83-Phe among Salmonella isolates.
Sequence and BLAST result of GyrA mutation at Asp87-Asp among Salmonella isolates.
Sequence and BLAST result of GyrA mutation for GAT-GAC (Asp-Asp) silent mutation among Salmonella isolates.
Discussion
Typhoid fever continues to be a global health problem and is endemic in developing countries including India. Worldwide about 12 to 33 million typhoid cases occurs each year [16]. In our study, 99.2% of S. Typhi showed resistance to nalidixic acid and 1/133 isolate of S. Typhi was found to be nalidixic acid sensitive; the same isolate was susceptible to the fluoroquinolone ciprofloxacin by MIC method. Only six isolates (5%) were sensitive to ciprofloxacin and most of the isolates showed intermediate resistant (74%, 99/133) to ciprofloxacin. Moreover, the ciprofloxacin MIC ranges (0.125- 0.5μg/ml) obtained for our study isolates were considered and reported by many authors as reduced susceptibility based on the clinical outcome of treatment with ciprofloxacin [4,17]. Our study showed better sensitivity for the first line drug ampicillin, which is similar with other reports [8,17].
The targets of fluoroquinolones are the two enzymes, DNA gyrase and topoisomerase IV, whose subunits are encoded respectively by gyrA and gyrB and the parC and parE genes [11]. The alteration caused by single point mutations within the QRDR of the DNA gyrase subunit gyrA gene leads to quinolone resistance (i.e. decreased susceptibility to ciprofloxacin) [17]. In Salmonella, the most commonly reported mutation leading to quinolone resistance was in Ser-83 and/or Asp-87 of gyrA gene [10,11,17,19]. Additional mutations may be required to attain high-level fluoroquinolone resistance [20,21]. Complete fluoroquinolone resistance in the Enterobacteriaceae usually results from two or more point mutations within the QRDRs of the DNA gyrase and topoisomerase IV genes [11].
An increase in the incidence of clinical Salmonella isolates with reduced susceptibility to the fluoroquinolones (i.e., ciprofloxacin MIC 0.125-0.5μg/ml) has been reported [22]. Though they are susceptible by in-vitro methods, treatment failures have been encountered when fluoroquinolones are used to treat infections caused by these strains. Several studies have shown mutation at Ser83-Phe in isolates which are resistant to quinolones [23,24]. When we analysed our S. Typhi isolates for gyrA gene mutations (125/133) the presence of mutation at Ser83 representing the substitution of Phe and a high level of resistance to nalidixic acid.
In the present study gyrA Ser83 mutation has been observed in 94% (125/133) of the clinical isolates. This report is in line with the previous reports of Renuka et al., who documented 84% of mutation in the NAR clinical isolates of Salmonella serovar with 92.1% of the isolates having gyrA Ser83 mutation [25]. 21.8% strains (29/133) shown both gyrA Ser83 and parC Trp106 mutations and all of them were resistant to both nalidixic acid as well as ciprofloxacin. Presence of ser83 mutation alone may cause nalidixic acid resistance and reduced susceptibility to ciprofloxacin hence the presence of these two mutations together could lead to high level resistant phenotypes.
The result showed mutation at Trp106 of parC gene in 21.8% (29/133) of S. Typhi. In general, mutation in parC is rare in gram-negative bacteria and usually arises after gyrA mutation, probably because in these organisms, gyrase rather than topoisomerase IV is the primary target of fluoroquinolones [17,26].
The results of present study showed Trp106 mutation of parC gene among the clinical S. Typhi isolates; which has not been much reported earlier. Previous studies showed parC gene mutation Thr57 detected in clinical isolates of Salmonella [27], parC gene mutations were observed mostly in animal isolates and very rarely in isolates from human origin [25,28]. As per our knowledge this study is the first to report of parC Trp106 mutation in clinical S. Typhi isolates.
The mutation status of the gyrA and parC genes and their MIC for nalidixic acid and ciprofloxacin of the Salmonella isolates were well correlated. The six Salmonella isolates which had mutation at Ser83-Phe and one Asp87-Asn of gyrA gene, showed nalidixic acid MIC of 256 μg/ml and ciprofloxacin MIC of 0.25-0.5 μg/ml. This study has found that the single most important factor contributing in reduced susceptibility to ciprofloxacin is mutation in the gyrase gene and topoisomerase IV gene.
Limitations
We have analyzed for mutations in the QRDRs of gyrA and parC and its association with fluoroquinolone resistance, whereas analysis of mutations in other targets (gyrB and parE) are yet to be done in detail.
Conclusion
In Salmonella, quinolone/fluoroquinolone resistance most commonly occurs due to mutations in the genes encoding DNA gyrase and topoisomerase IV. Uncontrolled use of the antibiotic could impose selective pressure on the organism, which may lead to the emergence of resistant strains. Salmonella isolates which showed intermediate resistance to fluoroquinolones could become highly resistant upon sequential accumulation of mutations in the target enzymes. Therefore, use of fluoroquinolones such as ciprofloxacin in the management of typhoid fever requires urgent review. Several studies suggested the reuse of former first line drugs such as ampicillin and chloramphenicol in the treatment of typhoid fever. Hence, reuse of the former first line drugs may reduce the pressure on Salmonella due to fluoroquinolone antibiotics. Continuous surveillance of the plasmid and chromosome of Salmonella species is also essential to alter treatment strategies aimed at maintaining the useful life of the few remaining antimicrobials available to treat enteric fever.
-, No growth