Traditionally GAS are identified as Gram positive cocci in chains showing beta haemolysis on 5% sheep blood agar and can be differentiated from other beta haemolytic streptococci (BHS) by bacitracin sensitivity, production of Pyrollidonyl Aryl Sulfatase (PYR) and Lancefield grouping based on the group specific cell wall polysaccharide antigen. Although many laboratories use bacitracin sensitivity as a presumptive identification test for GAS, it may give false positive results with Group C and Group G streptococci [5]. GAS identification by grouping is also not very reliable as there are antigenic cross reactions between other groups such as some strains of Streptococcus anginosus and Streptococcus dysgalactiae subsp. Equisimilis [6]. The recent advances in PCR technology targeting transcriptional regulator genes provide the most reliable and rapid method for detection of pathogenic bacteria [7]. Transcriptional regulators are specialized DNA binding proteins which play a crucial role in directing gene expression within bacteria for their adaptation and survival in different environmental conditions. Spy1258 is a putative transcriptional regulator gene (TetR/AcrR family) which is specific for S. pyogenes and can be used as a marker for its detection [8].
In the present study we compared the predictive values of routine phenotypic tests like bacitracin sensitivity, production of pyrollidonyl aryl sulfatase (PYR) and Lancefield grouping with spy1258 PCR for the identification of S. pyogenes.
Materials and Methods
This comparative analytical study was carried out in the Department of Microbiology, JIPMER, Puducherry, over a period of 18 months (1st November 2013 to 30th April 2015).
During this period a total of 34065 clinical samples were submitted for aerobic bacterial culture from various cases like pyoderma, abscess, tonsillitis, necrotizing fascitis, cellulitis, sepsis, etc. Two hundred and six beta haemolytic streptococci were obtained and included in the study based on preliminary tests like pattern of haemolysis on 5% sheep blood agar, Gram stain and catalase. For quality control S. pyogenes ATCC 19615 was included.
Identification Of Gas
Bacitracin Sensitivity [
5]
Test was performed using 0.04 units Bacitracin discs (Himedia Laboratories, Mumbai, India) as per standard protocol. After incubation, a zone of inhibition ≥ 15 mm was considered as sensitive.
PYR Test
The test was carried out using PYRA test (MIKROLATEST, Czech Republic). Briefly, 20 μl of distilled water was added onto the PYRA strip and the test organism was inoculated. The strips were incubated for 10 minutes at room temperature and 20 μl of reagent was added and the results were read after 2 minutes. The development of a red color indicates a positive test.
Latex Agglutination Assay [
6]
The Lancefield antigen was detected using STREP Test kit (Plasmatec, UK) according to the manufacturer’s instructions. A positive test was indicated by visible agglutination of the latex particles.
DNA Isolation
Genomic DNA was extracted by using mericon DNA Bacteria plus kit (Qiagen, Germany) according to the manufacturer’s instructions.
Spy1258 PCR
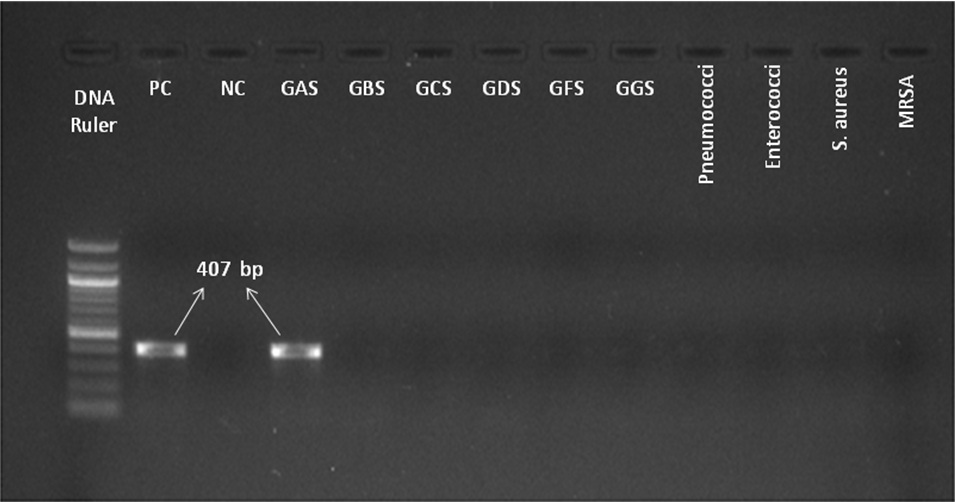
PCR was carried out in 15μl volume using Mastercycler nexus (Eppendorf, Germany). The reaction mixture contained 2x Taq PCR Smart Mix (Origin, India), 10 pmol each primers (forward primer: 5’-AAA GAC CGC CTT AAC CAC CT-3’ and reverse primer: 5’-TGG CAA GGT AAA CTT CTA AAG CA-3’) [8] and 10 ng sample DNA. The PCR cycling conditions: Initial heating at 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 20s, annealing at 55°C for 20s, extension at 72°C for 45s and final extension for 2 min after the last cycle. Electrophoresis of PCR product was carried out in 2% agarose gel and the presence of a 407 bp band indicate a positive test. Identity of the PCR product of one of the isolates was confirmed to be spy1258 gene by sequencing with Sanger’s method.
To check the specificity of spy1258 PCR we included Streptococcus pyogenes ATCC 19615, Staphylococcus aureus ATCC 25923 and clinical isolates of Group B,C,D,F,G Streptococci, Streptococcus pneumoniae, Enterococcus faecalis and MRSA [Table/Fig-1].
Gel Picture of spy1258 gene PCR product showing 407bp band specific for S. pyogenes. Lane 1- 100bp DNA ruler, PC- positive control, NC- negative control, GAS- Group A streptococcus, GBS- Group B streptococcus, GCS- Group C streptococcus, GDS- Group D streptococcus, GFS- Group F streptococcus, GGS- Group G streptococcus.
Statistical Analysis
Analysis was carried out using SPSS (Version 22.0). Categorical variables were presented in the form of percentage. The Sensitivity, Specificity, Positive Predictive Value (PPV) and Negative Predictive Values (NPV) were calculated for each test taking PCR as the gold standard. Kappa statistics was used to measure inter-observer agreement. The p-value <0.001 was considered statistically significant.
Results
Out of 206 BHS isolates tested, 176 (85.4%) were bacitracin sensitive, 174 (84.4%) were PYR positive, 177 showed positive agglutination with group A antiserum and 175 were PCR positive. One hundred and sixty isolates were positive for both phenotypic tests and PCR assay confirming as Streptococcus pyogenes and 17 isolates were negative for all tests suggestive of BHS other than Streptococcus pyogenes. The remaining twenty nine isolates had varying results with phenotypic tests as well as spy1258 PCR [Table/Fig-2].
Comparison between phenotypic tests and spy1258 PCR assay.
| Bacitracinsensitivity | Group A Latexagglutination | PYR test | Spy1258 PCR | No. of isolates(n= 206) |
|---|
| Sensitive | Positive | Positive | Positive | 160 |
| Resistant | Negative | Negative | Negative | 17 |
| Sensitive | Positive | Positive | Negative | 3 |
| Sensitive | Negative | Negative | Negative | 6 |
| Sensitive | Positive | Negative | Positive | 4 |
| Sensitive | Negative | Negative | Positive | 1 |
| Sensitive | Negative | Positive | Positive | 2 |
| Resistant | Negative | Positive | Negative | 2 |
| Resistant | Negative | Positive | Positive | 1 |
| Resistant | Positive | Positive | Positive | 4 |
| Resistant | Positive | Positive | Negative | 2 |
| Resistant | Positive | Negative | Positive | 3 |
| Resistant | Positive | Negative | Negative | 1 |
Nine of the bacitracin sensitive, seven each of PYR positive and group A antigen positive isolates were negative for spy1258 PCR. None of the 16 strains positive for other groups (B,C,D,F,G) were positive by the PCR assay. Four strains were positive for multiple groups along with group A, among them three were PCR positive. One isolate which was bacitracin resistant and negative for group antigen but PYR positive turned out to be PCR positive. Even though two of the three routine phenotypic tests failed to identify this particular isolate the spy1258 proved to be very sensitive in its detection.
The sensitivity, specificity, PPV and NPV of latex agglutination test was found to be slightly higher when compared with bacitracin sensitivity and PYR test. Kappa test showed substantial agreement between phenotypic tests and spy1258 PCR [Table/Fig-3].
GAS phenotypic identification in comparison to spy1258 PCR.
| Assay | Sensitivity(%) | Specificity(%) | PPV(%) | NPV(%) | Kappa value * |
|---|
| Bacitracin sensitivity | 95.42 | 70.96 | 94.88 | 73.33 | 0.673 |
| PYR test | 95.42 | 77.41 | 95.97 | 75.0 | 0.719 |
| Latex agglutination assay | 97.71 | 80.64 | 96.61 | 86.21 | 0.805 |
p-value of all <0.001 – statistically significant.
*Kappa Agreement: <0- Less than chance agreement, 0.01–0.20 Slight agreement, 0.21– 0.40 Fair agreement, 0.41–0.60 Moderate agreement, 0.61–0.80 Substantial agreement, 0.81–0.99 Almost perfect agreement
Discussion
Rapid and accurate identification of GAS is very important as mild skin or throat infections can lead to severe life threatening invasive conditions as well as post infectious immune mediated complications if left untreated [9,10].
The presumptive identification of Group A streptococci (GAS) is usually done by testing for sensitivity to bacitracin even though there are reports about the occurrence of bacitracin resistant strains [11]. Many laboratories use this as the sole test for diagnosing GAS infections [12]. In our study eight bacitracin resistant isolates were proved to be S. pyogenes by spy1258 PCR and nine bacitracin sensitive isolates turned out to be PCR negative. The sensitivity and specificity of this particular test was 95.42% and 70.96% respectively with a PPV of 94.88% and NPV of 73.33%. The results of our study indicate that the utility of bacitracin sensitivity test even as a preliminary test for screening GAS is debatable. In a recent study from Romania, out of the 460 isolates tested, bacitracin sensitivity was positive for only 92.75% of the isolates and 20.68% of the bacitracin resistant isolates (n=87) were further identified as Streptococcus pyogenes by latex agglutination test [13].
PYR test is considered to be specific for differentiating S. pyogenes from other BHS but it was reported that Streptococcus iniae and Streptococcus porcinus are also PYR positive [14]. In our study eight isolates which were PYR negative turned out to be S. pyogenes on spy1258 PCR. Five of the PYR positive isolates were PCR negative. The sensitivity and specificity of this particular test was 95.42% and 77.41% respectively with a PPV of 95.97% and NPV of 75%. The results indicate that this test should be supplemented with other tests for accurate identification.
The Lancefield system of serogrouping based on specific cell wall associated carbohydrates developed by Rebecca C Lancefield is useful in the identification of streptococci [15] and for long has been considered as the gold standard. Previous studies showed that it cannot be used as a single accurate test for identification, but should be supplemented with other tests like bacitracin sensitivity or PYR [6]. In our study when group A antigen positive isolates were subjected to PCR, 3.3% were negative and were identified as Streptococcus anginosus group. Another difficulty with latex agglutination test is lack of reproducibility of the results when repeated. This problem is compounded by the availability of different kits in the market with varying performance characteristics.
Recently MALDI-TOF has been developed as a tool for phenotypic identification of S. pyogenes with an identification accuracy ranging from 93.85% to 100% and it has been suggested as a rapid alternative to traditional biochemical tests for identification of S. pyogenes [16,17]. None of these studies included molecular based identification methods as control.
The major gene targets used for identification of S. pyogenes include spy1258, dnaseB, speB, and sof genes [7,18,19]. The latter three code for potent virulence factors. The sof gene code for serum opacity factor and is expressed by only 40-50% of GAS. DnaseB is not very specific as it is expressed in lower amounts by BHS other than Streptococcus pyogenes. SpeB gene is a chromosomally encoded structural gene which is present in almost all group A streptococci [20].
Spy1258 is a putative transcriptional regulator gene which is specific for S. pyogenes and may be involved in species-specific maintenance or adaptation [7]. It was noted that identical gene sequences of spy1258 were completely absent in other bacterial genomes available at GenBank. Many studies have used this particular gene for the rapid detection of S. pyogenes from various clinical samples. Al- Saadi et al., compared the sensitivity and specificity of spy1258 PCR and dnaseB PCR with conventional tests. Their sample size was very low; only 10 S. pyogenes isolates were included in the study. The sensitivity and specificity of spy1258 PCR was found to be 100% [21]. Another study reported that the sensitivity of spy1258 qPCR for detecting S. pyogenes directly from throat swabs was lower when compared with speB qPCR, which was attributed to a possible internal PCR inhibition [18]. Recently LAMP assay was developed and validated by Zhao et al., targeting spy1258 gene for rapid and accurate detection of S. pyogenes [22]. In the present study PCR for spy1258 proved superior to all the phenotypic tests as it was useful even in the isolates which were negative by the latter.
Applying PCR directly on clinical samples will further reduce the time required for identification of S. pyogenes and improve clinical outcomes especially in situations where early pathogen directed therapy is absolutely essential such as cases of rapidly progressing necrotizing fasciitis. This study is the first of its kind to determine the predictive values of routine phenotypic tests with molecular assay for S. pyogenes identification, incorporating a large number of clinical isolates.
Limitation
The limitations of the study include sequencing of only one representative isolate for confirmation as well as use of a single molecular target for identification of S. pyogenes.
Conclusion
In conclusion, clinical laboratories should not depend on bacitracin sensitivity as a single presumptive test for the routine identification of GAS but should use supplemental tests such as PYR test or latex agglutination test and for best results confirm by spy1258 PCR.
p-value of all <0.001 – statistically significant.
*Kappa Agreement: <0- Less than chance agreement, 0.01–0.20 Slight agreement, 0.21– 0.40 Fair agreement, 0.41–0.60 Moderate agreement, 0.61–0.80 Substantial agreement, 0.81–0.99 Almost perfect agreement