Periodontal disease consists of a group of infections that lead to inflammation of gingiva and destruction of periodontal tissues [1]. Histological evaluation of experimental gingivitis and chronic periodontitis provided considerable insight into the immunopathogenic process involved in the periodontal diseases [2]. A receptor molecule called “TOLL” was identified in Drosophilla species and it showed certain responses to infection or injury [3].
Toll Like Receptors are transmembrane proteins that recognize microbes once they have breached physical barriers and activate immune cell responses. The term TOLL originally referred to a cell surface receptor governing dorsal/ventral orientation in the early Drosophila larvae [4]. It was later found to also play a crucial part in antifungal defense, together with other antimicrobial peptides [5]. Sequencing of Drosophila genome revealed the existence of nine proteins belonging to the TOLL family [6].
All TLRs possess amino-terminal leucine-rich which are responsible for the recognition of Pathogen Associated Molecular Patterns (PAMPs) and also possess a carboxy-terminal Toll-Interleukin-1 (IL-1) Receptor (TIR) domain which is required for initiating intracellular signalling [7]. TLRs are primarily expressed by first line professional phagocytes such as macrophages, neutrophils and dendritic cells [8].
Over the last ten years considerable progress has been made regarding the discovery of TLRs, mutations and polymorphisms in TLRs, their signalling pathways and their potential role in the defence mechanism.
The aim and objective of the present study was to determine the corresponding level of expression of TLR2 and TLR4 in periodontal tissues of subjects with healthy gingiva and chronic periodontitis.
Materials and Methods
The present case control study was done at SRM Dental College and Hospital, Ramapuram, Chennai, India, for about six months. Tissue samples were obtained from 30 chronic periodontitis patients who were advised to undergo extraction of hopeless teeth that could not be saved by either Scaling and Root Planing (SRP) or flap surgery with or without bone grafting. Likewise, 30 tissue samples of subjects with healthy periodontium were collected who had undergone selective extraction for orthodontic treatment and dental caries. Specific tooth selection for the study was avoided as chronic periodontitis does not confine to a single or group of teeth.
The subjects with and without chronic periodontitis were diagnosed based on the criteria by American Academy of Periodontology classification; however the selected subjects did not suffer from any other systemic diseases. The patients who displayed clinical features of chronic periodontitis such as plaque and calculus accumulation, gingival inflammation and periodontal pockets were recorded by the OHI–S Index (John.C.Greene and Jack.R.Vermillion-1964) [9], Gingival Bleeding Index (Ainamo and Bay) [10] and Probing Pocket Depth respectively. The experimental group included inflammed gingival tissues with evidence of bleeding, local factors and pocket depth more than 6mm. The control group included periodontally healthy subjects with no evidence of bleeding, minimal local factors and pocket depth lesser than 4mm.
Patient with age group between 15 to 60 years of both genders were included in this study. Smokers, paan chewers, alcoholics, systemically compromised patients and patients who had undergone any surgery three months prior to sample collection were excluded from the study. The nature and design of the study was explained to the patients and written consent obtained for their participation.
These samples were transported to Madras Vertinary College after placing the sample in sterile eppendrof tubes containing saline. These tubes were kept in ice box during transportation. Samples were stored at -80° C till they were subjected to PCR analysis.
Isolation of Total Rna Using Trizol Method
Total cellular RNA was isolated from the tissue using TRIzol reagent (Invitrogen, USA) as per manufacturer’s protocol. Tissue samples were processed using liquid nitrogen. One ml of TRIzol was added to it immediately, homogenized and incubated at 15–30°C for five minutes. To that 200μl of chloroform was added and shaken vigorously. After incubation for 2–3 minutes at room temperature it was centrifuged at 12,000g for 5 minutes. Upper aqueous phase was transferred to a new tube and 500μl of isopropanol was added and kept at -20°C for two hours. After two hours it was centrifuged at 12,000 g for 10 minutes. Then the pellet was washed with 70% ethanol at 12,000g for 10 minutes and air dried. Finally, the pellet was resuspended in 20μl of RNase free water.
Synthesis of cDNA
The cDNA was synthesized from the extracted RNA as template using RevertAid Kit (Fermentas, USA). The reaction mixture was prepared as follows: 10μl of RNA was added to 2μl of random hexamer in clean sterile PCR tubes. The tubes were incubated at 70°C for 5 minutes and immediately snap chilled. Then, 4μl of reaction buffer, 2μl of dNTPs, 1μl of MuMLV reverse transcriptase enzyme and 1μl of RNase inhibitor were added. Final volume of 20μl reaction was run in thermal cycler with the following cycle condition 25°C–10 min, 42°C–1hr, 72°C–5 min and 4°C-holding.
The cDNA was then stored at -70° C until further use.
Reverse Transcriptase – PCR (RT-PCR) Using TLR Primers:
RT–PCR was done using synthesized cDNA (as template) and TLR primers. The following primer sets were used for real-time PCR assays:
TLR2 sense: 5’-TTTCA CTGCTTTCAACTGGTA-3’, TLR2 anti-sense: 5’-TGGAGAGG CTGATGATGAC-3’, TLR4 sense: 5’-CGATTC CATTGCTTCTTG-3’ and TLR4 anti-sense: 5’-GCTCAGGTCCAG GTTCTT-3’. Housekeeping gene (β–lactin) was used as an internal control. PCR was done in a thermal cycler with initial denaturation of 94°C for 5min, denaturation of 94°C for 30sec, annealing of 59°C for 30sec, extension of 72° C for 1 min, final extension of 72° C for 7min and holding of 4°C
After 35 cycles the amplified PCR products were stored in 20°C until further use.
Agarose Gel Electrophoresis
Agarose gel (1.5%) was prepared by adding 0.375 gm of agarose in 25ml of TAE buffer. Agarose was melted and casted on a gel tray after adding 3μl of ethidium bromide to it. After solidification the gel was placed on electrophoresis tank containing 1xTAE buffer. The PCR mix was then loaded to each well along with 100bp molecular marker (Banglore Genei). The agarose gel was run with a constant 80V for 20-30 min and documented using gel documentation unit [Table/Fig-1,2]. Results were thus recorded and statistically analyzed.
(a) Expression of TLR2 with 100 bp DNA ladder in lane 7. (b) Expression of TLR2 with 100 bp DNA ladder in lane 8. (c) Expression of TLR2 with 100 bp DNA ladder in lane 8. (d) Expression of TLR2 with 100 bp DNA ladder in lane 8. (e) Expression of TLR2 with 100 bp DNA ladder in lane 8. (f) Expression of TLR2 with 100 bp DNA ladder in lane 8.
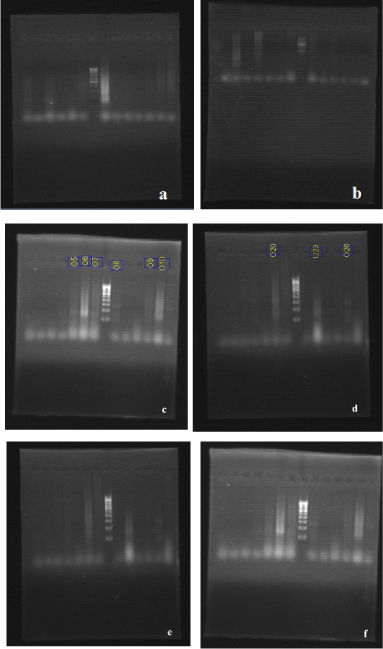
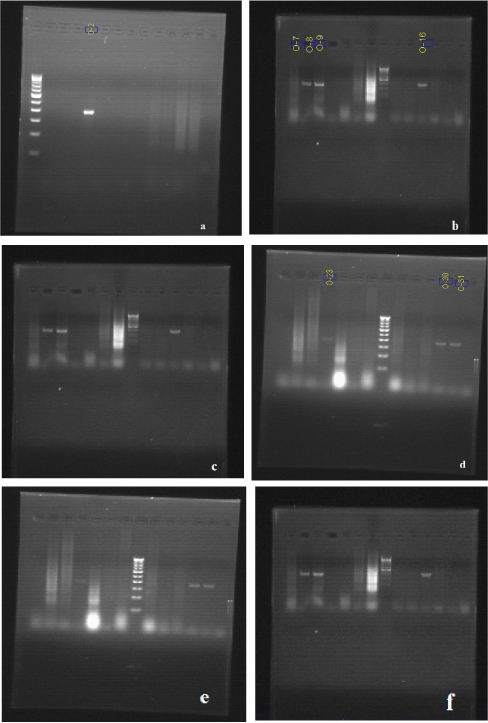
(a) Expression of TLR4 with 100 bp DNA ladder in lane 1. (b) Expression of TLR4 with 100 bp DNA ladder in lane 8. (c) Expression of TLR4 with 100 bp DNA ladder in lane 8. (d) Expression of TLR4 with 100 bp DNA ladder in lane 8. (e) Expression of TLR4 with 100 bp DNA ladder in lane 8. (f) Expression of TLR4 with 100 bp DNA ladder in lane 8.
Results
[Table/Fig-3] shows that the mean Gingival Bleeding, Oral Hygiene Index-S and Pocket Depth were highly significant in the Experimental Group than the control group which implies that the signs of clinical inflammation were seen more in the experimental group.
Comparison of mean values between control and experimental groups.
| Variables | Experimental Group | Control Group | p-value |
|---|
| Mean ± S.D. | Mean ± S.D. |
|---|
| Gingival Bleeding | 97.0 ± 12.7 | 78.3 ± 36.6 | 0.009 (Sig)$ |
| Oral Hygiene Index-S | 2.303 ± 0.3624 | 1.317 ± 0.5730 | 0.000(Sig)$ |
| Pocket Depth | 6.90 ± 2.440 | 1.80 ± 0.714 | 0.000(Sig)$ |
$ Mann-Whitney U test was used to calculate the p-value.
It is inferred from [Table/Fig-4] that there was no significant association between TLR2 and study groups (p=0.085) whereas there was a significant (p =0.004) association between TLR4 and the study groups. This shows that the TLR2 distribution is similar in both study groups and the proportion of positivity for TLR4 in experimental group was significantly higher than the control group. Thus, in subjects with chronic periodontitis (experimental group), the levels of TLR4 were identified more than the control group.
Association of TLR2 and TLR4 with study groups.
| Variables | Response | Control Group | Experimental Group | p–value |
|---|
| No. | % | No. | % |
|---|
| TLR2 | Negative | 29 | 96.7 | 25 | 83.3 | 0.85 (NS) |
| Positive | 1 | 3.3 | 5 | 16.7 |
| TLR4 | Negative | 27 | 90.0 | 17 | 56.7 | 0.004 (Sig)$ |
| Positive | 3 | 10.0 | 13 | 43.3 |
$ Pearson Chi-Square test (2-tailed) was used to calculate the p-value
Discussion
The pathogenesis of periodontal disease is mainly a result of host response to microbial induced tissue destruction. Any injury or insult to the tissues will initiate the host immune and inflammatory processes. The innate immune system is activated by the participation of pattern recognition receptors (PRR) [11]. The PRR such as TLRs are signal molecules essential for the cellular response to bacterial cell wall components [11]. These microbial structures are referred to PAMPs and include bacterial lipopolysaccharides, lipoprotein, peptidoglycans, bacterial DNA and double stranded RNA [8]. TLR signaling is thought to be one of the most important pathways initiating the onset of periodontitis [12].
Ten human TLRs have been identified till now and among all, TLR 2 and 4 play a pivotal role in periodontal disease [13]. Human gingival fibroblasts and periodontal ligament fibroblasts are representative constituents of the periodontal tissues which produce various inflammatory cytokines along with the TLR2 and TLR4 [3].
The results showed that both TLR2 and TLR4 were expressed in the control group and the experimental group. This expression in the control group supports the literature of its presence in the healthy gingival [14]. But TLR4 in the experimental group was expressed more with a significant p-value (0.004) than TLR2 in both groups and TLR4 in the control group (not significant) [Table/Fig-4]. This experimental group showed a significant increase in mean gingival bleeding, oral hygiene index-S and pocket depth suggesting more clinical signs of inflammation compared to the control group [Table/Fig-3] thus proving that the elevated levels of TLR4 in chronic periodontitis correlates with the clinical parameters observed.
The gram negative organisms and their products such as Porphyromonas gingivalis and lipopolysaccharides responsible for chronic periodontitis are well recognised by TLR-4 [15,16]. Studies done by Daisuke Abe et al., states that the TLR4 signalling complex (lymphocyte antigen 96) on binding to the extracellular domain of TLR produces cytokines and chemokines which facilitate LPS-mediated NF-kB activation [12]. Hence, the TLR4 is expressed more in chronic periodontitis corelating with the results of this study.
TLR2 is unique in that it heterodimerizes with the signalling partner TLR1 or TLR6 for detecting and responding to microbial cell wall components such as lipoteichoic acid, peptidoglycan and lipoprotein or lipopeptides [8]. Studies done by Sarah et al., has shown that the expression of TLR2 in gingivitis samples were elevated due to the fact that TLR2 recognizes the above mentioned PAMPS which is majorly seen in gingivitis [17]. Moreover, studies done by George Hajishengallis et al., had also shown that P.gingivalis acting through TLR2 may stimulate a different inside-out signalling pathway from that activated by LPS acting through TLR4 which is seen in chronic periodontitis [18]. This further supports the fact that TLR2 will be increased in gingivitis than in chronic periodontitis samples.
On thorough literature research, the results of similar studies were tabulated for comparison with the results of the present study [Table/Fig-5].
Review of similar studies and their respective results.
| AUTHOR & YEAR | STUDIES | RESULTS |
|---|
| Beklen A et al., (2014) | The function of TLR4 in interferon gamma or interleukin-13 exposed and lipopolysaccharide stimulated gingival epithelial cell cultures. | Periodontitis tissue samples showed increased TLR4 levels [19]. |
| D’souza RS et al.,(2013) | Analysis of expression and localization of TLR-2 by immunofluorescent technique in healthy and inflammed oral tissues. | The levels of expression of TLR2 is increased in chronic periodontitis and seen higher in the epithelial cells than in the connective tissue cells [20]. |
| Wara-aswapati N et al., (2013) | Induction of toll-like receptor expression by Porphyromonas gingivalis. | The levels of TLR2 and TLR4 were significantly increased in periodontitis patients [21]. |
| Muthukuru M et al., (2005) | Oral mucosal endotoxin tolerance induction in chronic periodontitis. | TLR4 cells increases in subjects with chronic periodontitis than TLR2 cells [22]. |
| Mori Y et al., (2003) | Immunohistochemical localization of Toll-like receptor 2 and 4 in gingival tissue from patients with periodontitis. | The expression of TLR2 and TLR4 positive cells were increased in mild and severe gingivitis tissue samples respectively [23]. |
| Wang PL et al., (2003) | DNA micro array analysis of human gingival fibroblasts from healthy and inflammatory gingival tissues. | Increased TLR2 levels in human gingival fibroblasts of inflamed gingiva [24]. |
| Schwandner R et al., (1999) | Peptidoglycan and lipoteichoic acid- induced cell activation is mediated by TLR 2. | Identified TLR2 as a signal transducer for peptidoglycans and lipotechoic acid in addition to lipopolysaccharides [25]. |
The present study results are in correlation with the results of most of the similiar studies tabulated, excluding the contrasting result from the study done by D’souza RS et al., where in the TLR2 levels were not evaluated in comparison with TLR4 levels [20]. We could not compare our present study results with studies done by Mori Y et al., and Wang PL et al., as our study group did not include gingivitis patients [23, 24]. This limits the scope of TLR2 and TLR4 analysis in various stages of gingivitis. This can be viewed as a limitation of this present study.
To summarize, this present study reveals the active participation of TLR4 than TLR2 in the disease process of subjects consisting of chronic periodontitis. Hence, it can be clinically implied to diagnose chronic periodontitis at its early stages and reduce the inflammatory spread by treating it at the earliest. In the future, TLR manipulation can also be done for modifying the periodontal disease process.
Conclusion
The present study concludes that TLRs act as a good indicator of inflammatory activity helping us to understand the periodontal disease and its progression. Unlike other traditional methods like probing depth, it evaluates the status of present disease activity in an inflammatory condition. In future, further studies have to be done with these scientific findings to confirm TLRs as a diagnostic biomarker for periodontal diseases.
$ Mann-Whitney U test was used to calculate the p-value.
$ Pearson Chi-Square test (2-tailed) was used to calculate the p-value