Majority of Urinary tract infections (UTI) encountered in clinical practice are simple acute uncomplicated cystitis [1]. Escherichia coli and Klebsiella pneumoniae are the frequent uropathogens that play important roles in UTI. Production of Extended spectrum beta lactamase (ESBL) by E.coli and K.pneumoniae are associated with co-resistance to fluoroquinolones, co-trimoxazole, aminoglycosides which are frequently used in treating UTI [2].
Nitrofurans are group of compounds that are characterized by presence of nitro groups on nitro aromatic ring. Nitrofurantoin is an underused oral antimicrobial agent, rapidly absorbed and excreted in urine to generate high therapeutic concentrations [3]. It has been used for treating nosocomial and community-acquired lower urinary tract infections. Though nitrofurantoin has been in clinical practice since 1952 with an extensive spectrum of activity, there was no upward trend in acquired resistance to this drug. In spite of various merits of nitrofurantoin, many clinicians are familiar with the side effects of the drug than being aware of its benefits.
Nitrofurantoin can act as a proper antibiotic only after it is reduced by nitro reductase enzymes.
Nitrofurantoin resistance has been recorded with only genes of oxygen insensitive nitro reductase enzymes. Genetically, step wise mutations of different nitro reducing activities present in E. coli are identified as the reason behind the progressive increase in resistance patterns [3]. These genes are mapped as nfsA and nfs B genes.
This study was planned to define the resistance pattern of nitrofurantoin in uropathogens and to identify the genes responsible for the same and the type of mutations involved. Moreover, the in vitro efficacy of nitrofurantoin in different types of multi drug resistant uropathogens was also analysed.
Materials and Methods
This study was conducted in a tertiary care hospital for a period of six months (November 2013 – April 2014) which caters to a total of 1200 beds.
Institutional ethics committee approval was obtained (CSP – MED/13/OCT/09/101). A total of 115 isolates with significant bacteriuria were included in the study. Among 115, ESBL producers; carbapenamase producers and non-ESBL producers were collected. Organisms were identified up to species level by conventional methods [4] and Microscan Walkaway system 96.
1. Antibiotic susceptibility testing: Antibiotic susceptibility testing was done by Kirby- Bauer disk diffusion method [5] according to CLSI guidelines 2013 [6] by using disks procured from Hi Media Laboratories, Mumbai. The antibiotics namely Ampicillin (10μg), Cephalexin (30μg), Cefuroxime (30μg), Cefotaxime (30μg), Ceftazidime (30μg), Cefaperazone-sulbactam (75/10μg), Piperacillin -tazobactam (100/10μg), Amikacin (30μg), Trimethoprim/sulfamethoxazole (1.25/23.75μg), Imipenem (10μg), Meropenem (10μg), Ertapenem (10μg), Norfloxacin (10μg), Ofloxacin (5μg) and Polymyxin B (300U) were tested.
Escherichia coli ATCC 25922; Klebsiella pneumoniae ATCC 700603 were used as quality control strains.
2. Susceptibility to Nitrofurantoin: Nitrofurantoin (300μg) was used for disk diffusion test. A zone size of >17mm was considered sensitive, <14mm as resistant and 15-16mm as intermediate.
3. Screening for ESBL and Carbapenamase production: ESBL production were tested with resistance to third generation cephalosporins and susceptible to beta lactamase inhibitors. Carbapenemase production tested with resistance to imipenem or meropenem.
4. Minimal inhibitory concentration (MIC): MIC to Nitrofurantoin was done by E-strip obtained from Himedia, Mumbai. All study isolates with reduced susceptibility to Nitrofurantoin were selected for MIC. Resistant isolates were identified with MIC >128 μg/ml, intermediate - 64 μg/ml and sensitive - <32 μg/ml.
5. Molecular Characterisation:DNA Extraction: DNA isolation was performed in all the clinical isolates and standard strains by Phenol: Chloroform method. Concisely, cell lysis was done by taking 400μl of lysis buffer in which a loopful of bacteria was suspended along with sterile glass beads in a 1.5ml centrifuge tube and vortexed for 2 minutes. Deproteinisation was done by centrifuging a mixture of equal volume of Phenol: Chloroform at 10,000 rpm for 10 minutes. After removal of the aqueous layer, 500μl of chloroform was added to the supernatant and the process was thus repeated once. Cold isopropanol in equal amounts was used to precipitate the DNA and was further centrifuged and washed with 70% ethanol to make a pellet, which was resuspended in 40μl of TE buffer and stored at -20° C until use.
Polymerase Chain Reaction:Escherichia coli strains with resistant and intermediate MIC were subjected to conventional PCR for detection of nitroreductase nfsA gene. Susceptible E.coli were used as control. Primers were obtained from Sandegren et al., [3] 2008.
Fwd 5’ - ATTTTCTCGGCCAGAAGTGC - 3’,
Rev 5’ - AGAATTTCAACCAGGTGACC - 3’
Fwd1 5’ - TTTTCTCGGTGTTTTGCTCA-3’
Rev1 5’- GCTGTATAGCGGCTTCACG-3’
PCR Assay: The master mix was prepared by adding 25μl of PCR mix (GeNei, Bangalore), 1μl of forward and reverse primer respectively, 1μl of template DNA and the volume was made up to 50μl with sterile nuclease free water. Then the reaction mix was kept in the thermocycler. The procedure included initial denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 seconds, followed by annealing at 56°C for 30 seconds and extension at 72°C for 30 seconds and lastly by final extension at 72°C for 5 minutes.
Gel Electrophoresis: PCR products were electrophoresed in 1.5% agarose gel, stained with Ethidium bromide (0.5μg/ml) and visualized under UV light and photographed.
Gene Sequencing: The PCR products were then sent for gene sequencing to Scigenom Labs, Cochin, Kerala, India.
BLAST Analysis and Multiple Sequence Alignment: The sequence was then used for a nucleotide - nucleotide search using the BLAST algorithm at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/). BLAST hits more than 99% were considered. Representative sequence from sensitive, intermediate and resistant strains were taken and pair wise alignment was performed using ClustalW http://www.ebi.ac.uk/Tools/msa/clustalo/). Results were then compared.
Results
Of the total 115 clinical isolates, 70 E.coli and 45 Klebsiella pneumoniae were included in the study. This included 63 ESBL producers, 45 carbapenem resistant isolates and 7 nitrofurantoin susceptible strains which were non-ESBL and carbapenem susceptible.
1. Disk diffusion method: In E.coli ESBL production was noted in 64.2% (n=45) and 40% (n=18) of K.pneumoniae isolates. Resistance to carbapenems was seen with 32.8% (n=23) of E.coli and 48.8% (n=22) of Klebsiella pneumoniae.
Among nitrofurantoin susceptible strains 58% (n=37) were ESBL producers and 37% (n=17) were carbapenems resistant strains.
In E.coli 50% (n= 35) were susceptible, 11.4% (n=8) intermediate and 38.5% (n=27) were resistant to nitrofurantoin whereas in Klebsiella pneumoniae 13.3% (n= 6) were susceptible; 22.2% (n=10) were intermediate and 64.4% (n=29) were resistant to nitrofurantoin by disk diffusion method [Table/Fig-1].
Table showing number of isolates that are resistant to various classes of antimicrobial agents and the percentage of nitrofurantoin susceptibility among the resistant strains.
| Antimicrobial agent | Escherichia coli | Klebsiella pneumoniae |
|---|
| Resistant | Nit susceptible | Resistant | Nit susceptible |
|---|
| Co-trimoxazole | 75.7% (n=53) | 40% (n=28) | 68.8% (n=31) | 11%(n=5) |
| Quinolones | 88.5% (n=62) | 42.8% (n=30) | 55.5% (n=25) | 4.4%(n=2) |
| β Lactam inhibitors | 28.5% (n=20) | 11.4% (n=8) | 42% (n=19) | 6.6% (n=3) |
| Aminoglycosides | 12.8% (n=9) | 4.28% (n=3) | 37.7% (n=17) | 6.6%(n=3) |
| Polymyxin B | 4.28% (n=3) | n=0 | 2.2% (n=1) | n=0 |
2. Minimal inhibitory concentration: MIC to Nitrofurantoin ranged from 4 to >512 μg/ml. MIC of nitrofurantoin resistant E.coli and K. pneumoniae strains by disk diffusion ranged from 8 to >512 μg/ml and intermediate strains ranged from 16 to 128 μg/ml by E- strip method [Table/Fig-2].
Table showing number of isolates that are susceptible, intermediate and resistant to nitrofurantoin by E – strip method (MIC).
| Nitrofurantoin | E.coli | Klebsiella pneumoniae |
|---|
| Resistant | 12(17.1%) | 23(51%) |
| Intermediate | 9(12.9%) | 9(20%) |
| Sensitive | 49(70%) | 13(29%) |
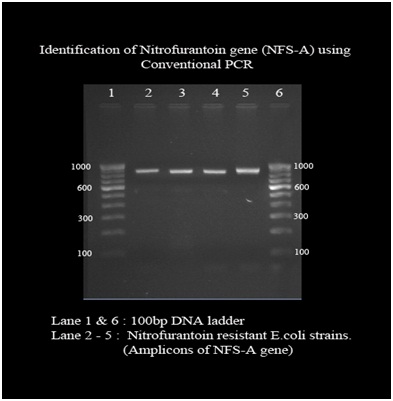
3. Polymerase chain reaction: Conventional PCR detected nfsA nitroreductase gene in 64 out of 70 E.coli strains with the band size of approximately 900bp. Out of 70 isolates, all the sensitive strains produced band of same size whereas only 15 out of 21 resistant and intermediate isolates produced amplicon. Other six resistant isolates failed to amplify in the PCR reaction [Table/Fig-3].
Gel electrophoresis picture under UV light where the resistant E.coli isolates showing amplicon of 900 base pairs.
4. Gene sequencing: Representative strains of sensitive, intermediate and resistant isolates were sent for sequencing. The sequence was then used for a nucleotide - nucleotide search using the BLAST algorithm at the NCBI website (http://www.ncbi.nlm.nih.gov/BLAST/). The BLAST hits showed 99% identity to the nfsA gene from the E.coli genome which is available in the NCBI database.
Sensitive, Intermediate and Resistant strains were taken and pair wise alignment was performed using online tool, Clustal W omega at (http://www.ebi.ac.uk/Tools/msa/clustalo/).
On comparing all three sequences, in resistant strain there was one insertion mutation seen in which four new base pair were added. There were 20 substitution mutations observed in both intermediate and resistant isolates [Table/Fig-4].
CLUSTAL O(1.2.1) multiple sequence alignment showing susceptible, resistant and intermediate sequences and the mutations observed were highlighted.
S- susceptible strain; R- resistant; I- intermediate
ATGC – Mutation in Both Resistant and Intermediate Strains
ATGC – Mutation in Intermediate strains
ATGC – Insertion Mutation in Resistant Strains

Sequenced strain was submitted to Genbank and was provided a GenBank accession number for the nucleotide sequence KR 296984.
Discussion
The spread of antimicrobial resistance among bacterial pathogens has emerged as an important challenge for the medical community. In spite of widespread use of nitrofurantoin for more than six decades, there has been practically no acquired resistance to nitrofurantoin. Karlowsky et al., has revealed a minimal annual difference of about 0.1% for nitrofurantoin during 1998-2001 with various urinary antimicrobials [7].
Debasis Biswas et al., and Naveen et al., have demonstrated that nitrofurantoin can be effectively used in E.coli and K.pneumoniae [8,9]. Likewise a study by Naveen et al., reported that E. coli isolates were mostly susceptible to amikacin (80.4%), followed by ceftriaxone (60.9%), gentamicin (36.5%), nitrofurantoin (34.1%), and norfloxacin (19.5%) [9]. K. pneumoniae isolates were mostly susceptible to amikacin (68%), followed by ceftriaxone (60%), gentamicin (40%), nitrofurantoin (32%), and norfloxacin (16%). In this study it was observed that 65 % of ESBL producers and 51% of carbapenemase producers were susceptible to nitrofurantoin by minimal inhibitory concentration. Whereas only 22% of ESBL producers and 31% of carbapenamase producers were susceptible to quinolones and 22% of ESBL producers and 37.7% of carbapenamase producers were susceptible to co-trimoxazole. This study demonstrated Nitrofurantoin had a higher susceptibility rate when compared to quinolones and cotrimoxazole in ESBL producers and carbapenamase producers. In a study by Puerto et al., among 115 clinical isolates of E. coli ESBL producers, 71.3% were susceptible to nitrofurantoin [10]. Also, Chen et al., completed their study by commenting that nitrofurantoin, possibly will be an alternative in the treatment of ESBL-producing E. coli-related uncomplicated UTI [11].
Cotrimoxazole and quinolones were among the few oral therapeutic options for ESBL-producing isolates [2]. But in this study, co-trimoxazole was the least active antimicrobial agent against ESBL producing Klebsiella isolates (11%) and quinolones were least active among ESBL- E.coli isolates (7%). Ko et al., showed that the resistance rates for trimethoprim-sulfamethoxazole among E.coli are growing accompanied with ciprofloxacin resistance and ESBL production [12]. Debasis Biswas et al., in their study had reported that nitrofurantoin and amikacin recorded the least resistance for E.coli [8]. nitrofurantoin was found to be the least effective against the amikacin resistant strains. Eight isolates were resistant to both Nitrofurantoin and Amikacin. In this study 15 (13%) isolates were resistant to both the agents.
Frank resistance of total 115 isolates to nitrofurantoin in our study was 30%. While resistance to nitrofurantoin was almost 10.2% in a study by Ehsan Valavi in 2013, indicating that nitrofurantoin could be a suitable choice for the treatment and prophylaxis of cystitis [13]. A study by Huang and Stafford suggests that many ambulatory care physicians already prescribe nitrofurantoin and fluroquinolones to treat urinary tract infections in women [14]. Proven that there is occurring of switch in therapy, clarity with concern to efficacy of nitrofurantoin among uropathogens that are resistant to other classes of antimicrobials is important and of benefit to clinicians.
In the present study, all the E.coli isolates produced PCR amplicon for nfsA gene approximately 900bp in size except 6 E.coli nitrofurantoin resistant strains. In all the resistant strains MIC values were found to be >96 mcg/dL. Earlier, Sandegren et al., had reported that all single step in vitro mutants had mutations in nfsA gene while some strains had no PCR product with any of primer combinations [3]. The same was experienced in our study, as six E.coli isolates with similar resistant pattern for the nitrofurantoin drug did not give amplicon. The possible explanation for non-existence of PCR product might be due to incidence of larger relocations or deleterious mutation in the primer binding area in the nfsA gene [3]. The mutation in primer binding site or absence of the same due to mutation could have affected PCR amplification and resulted in absence of PCR product. Representative isolates from sensitive, intermediate and resistant strains were sequenced and compared by multiple sequence alignment. The resistant isolates had insertional mutation of four nucleotides when comparing with the sensitive and intermediate strain sequences, which eventually alters the triplet codon in the translation process of protein synthesis, thus synthesizing biologically inactive nitroreductase enzyme. The inactive enzyme cannot reduce nitrofurantoin which inhibit the production of toxic byproducts. Among the 20 mutations isolated in the study by J Whiteway et al., in 1998, in the nfsA genes of the nitrofuran derivative resistant mutants, 13 (65%) were derived from the insertion of one of six IS elements [15].
Apart from the insertional mutation we also encountered substitutional mutation, in which the nitrogen bases were replaced by another. A total of 8 substitutional mutations were found in the resistant strains compared to sensitive strain [Table/Fig-4]. The mutation caused transition of A→G, T→ C, C→T, and G→A. Since these are the point mutations, neither does it not affect the codon position nor the end product, hence these mutations will not aid in resistant property of the organism. All the point mutation and nitrogen base transition present in the resistant strain sequences were also present in all the intermediate strains. Apart from those eight mutation, in intermediate strain we found four additional point mutations which makes the total mutation count to 12 [Table/Fig-4]. Those nitrogen base transition were T→A, G→T and A→C. As this transition did not alter the protein sequence heavily, the organism could produce the nitroreductase enzyme without defect and hence organism becomes susceptible to the drug. Other than these 12 mutations, we did not find any deleterious mutations in the intermediate strains.
Surprisingly, resistance to nitrofurantoin remains minimal and may be related to the fact that it has multiple mechanisms of action hence may demand the organisms to develop more than a single mutation to concur resistance. Similar review was mentioned by Shakti L et al., that to acquire resistance to nitrofurantoin by an isolate it has to undergo step wise mutations [16]. The finding in our study suggests that the strains underwent several number of mutations during the infection spread, but not all the mutations can cause alteration in protein structure. Insertional mutation found in resistant strain is the cause of defective enzyme production thereby substantiating the resistance to the drug. Although multiple number of substitutional mutations are encountered they are merely silent mutations which just replaces the nucleotide bases alone, thereby no alterations in the protein product. Identification of nfsA gene using conventional PCR and gene sequencing could help to screen the mutation in resistant strains and the cause of mutation. Combination of conventional drug resistant identification and gene analysis would help in prompt identification of resistant strains and effective treatment.
Limitation
In this study nfsA gene has been analysed only in E.coli isolates but not in Klebsiella pneumoniae. Molecular study of nfsB gene is not involved in the present study which has to be determined in near future.
Conclusion
Nitrofurantoin being an oral antibiotic, its usage in ESBL producers and carbapenamase producers is still warranted with susceptibility rate reaching around 46% in ESBL and almost 15% in carbapenamase producers. The urinary antibiotics including nitrofurantoin and fluoroquinolones especially norfloxacin, which are used in treatment of urinary tract infections, achieve high therapeutic concentrations in urinary bladder and the likelihood of their in-vitro resistance readings in the laboratory may not necessarily translate into treatment failure. Also, on the other hand, patients with afebrile community-acquired UTI can be treated more conventionally with oral antibiotics such as nitrofurantoin, particularly in view of the very low resistance of the most common pathogen E. coli to nitrofurantoin study. Likewise, restricted use of antibiotics and combination therapy may limit the increasing pattern of antibiotic resistance.