Introduction
Aptamer are a new class of agents that can be used for both diagnostic and therapeutic purposes. They are synthetic single strand (ss) DNA or RNA molecules. They are selected against target molecules by an iterative process known as SELEX (Systematic Evolution of Ligands by Exponential Enrichment) [1], which was developed in 1990. Due to its various advantages, aptamers are regarded as promising alternatives to antibodies. Aptamers could be massively synthesized using in vitro techniques; its production is cost effective and animal free in nature. Aptamers bind specific ligands with high affinity and selectivity. They are more robust at elevated temperatures and thermal denaturation is reversible [2]. In last few years aptamers have been widely used in therapeutics. Macugen was the first FDA approved drug that was used against macular degeneracy disease [3]. Aptamers are also being devolved that can be used in clot buster, cancer therapy, autoantibodies, diabetes etc [3]. Companies such as NOXXON, Anisoma and other are doing cutting edge research on aptamers to be used as drug.
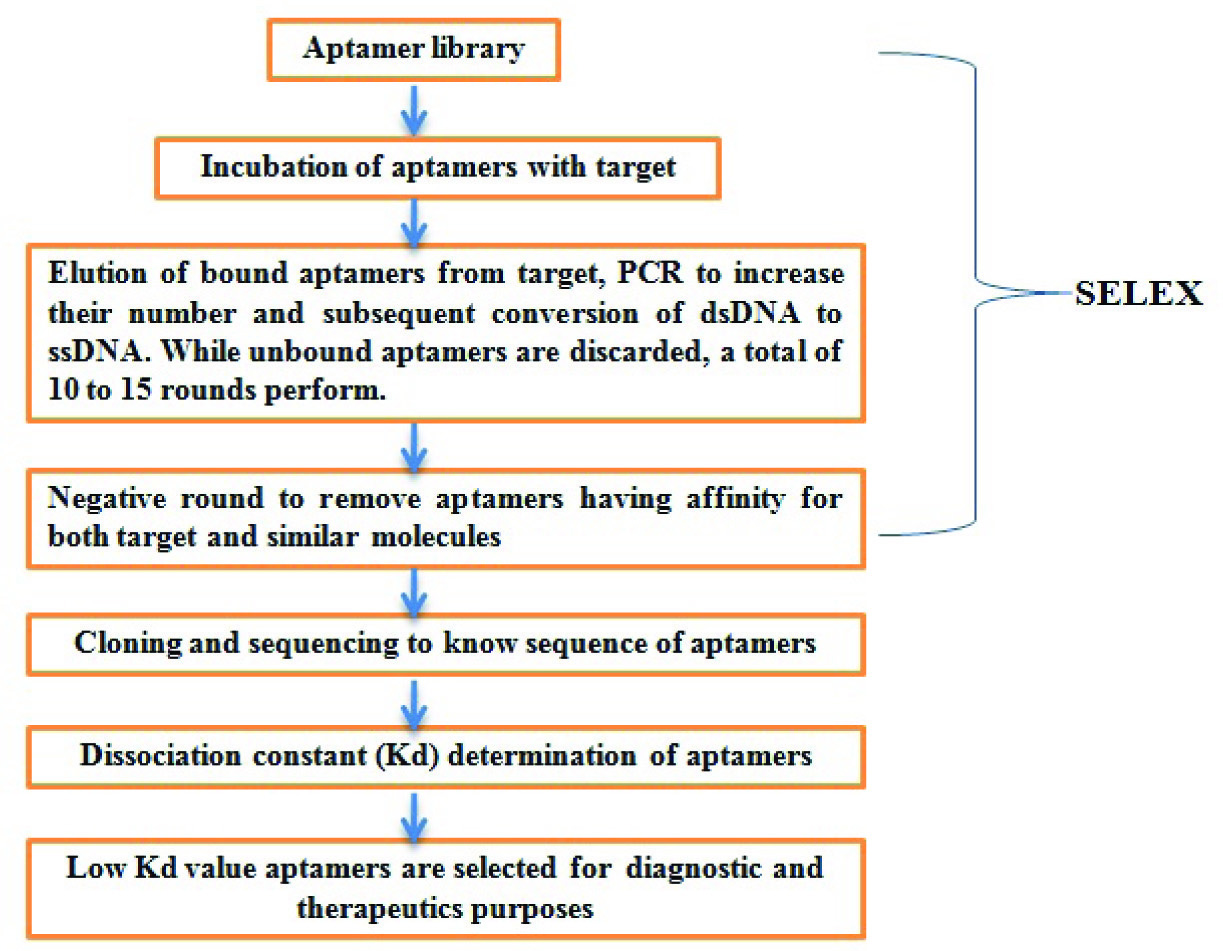
Aptamers and SELEX
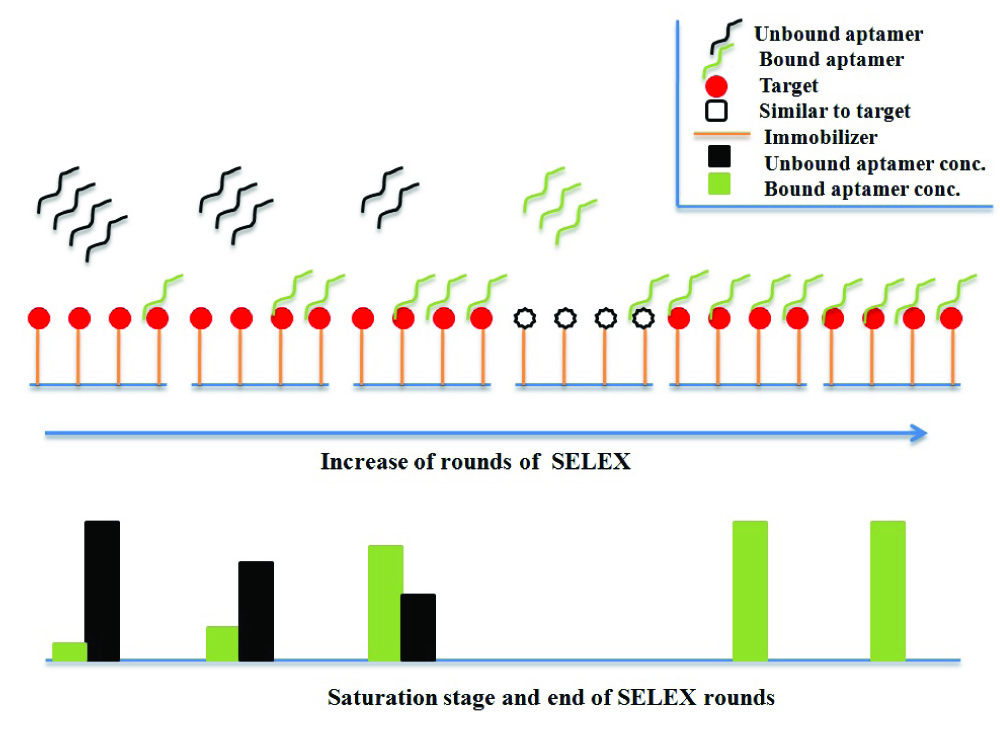
Aptamers are synthetic DNA or RNA molecules that are custom made [4]. Artificial DNA has length in the range of 60 to 100 nucleotides. The 5’ and 3’ end of aptamer contains bases which are common in all ssDNA or aptamers and their ends can be 15 to 18 bases long. Rest of the region is middle region known as random region, contains bases at different positions. These unique DNA molecules together form DNA or aptamer library. A typical library contains more than 1015 different ssDNA molecules. Aptamer library is required to start SELEX process for aptamer selection against target molecule. SELEX is an iterative process where DNA library is incubated with target. Due to randomness in library, some of the aptamer binds to target while rest are discarded. The bond between the aptamer and target is broken down by using urea, EDTA at high temperature. Eluted aptamers are amplified by Polymerase Chain Reaction (PCR). This converts ssDNA to double strand (ds)DNA which is converted back to ssDNA for next round of SELEX. This process is known as one round of SELEX. A total of 10 to 15 rounds of SELEX are performed to get aptamers having very high affinity for target. During SELEX, similar but not identical molecules to target are also incubated with aptamers to discard any aptamer having affinity for both target and similar molecules. This process is known as negative or counter selection round [5]. Selected aptamers can be used for diagnostic as well as therapeutic purposes [Table/Fig-1,2].
Application of Aptamers in Therapeutics
The first SELEX experiment was carried out by Tuerk and Gold in 1990, when they selected RNA aptamer against bacteriophage T4 DNA polymerase [1]. In literature we can find large number of aptamers specific against wide variety of targets [6–13].
Aptamers Against Age-related Macular Degeneration (AMD)
i) Macugen- It is an RNA aptamer that consists of 28 nucleotides and also known as pegaptanib. Macugen was the first FDA (in 2004) approved drug used in treatment of wet AMD (age-related macular degeneration) [14,15]. It was initially developed by NeXstar pharmaceuticals and in 2000 license was given to EyeTech Company (now OSI Pharmaceuticals) for late stage development and marketing in the United States. From outside of US it is marketed by Pfizer. Its molecular formula is C294H342F13N107Na28O188P28(C2H4O)2n, (n=900), M.W is 50 kDa and biological half time is 10 days. Macular degeneration is a disease of eyes where excessive leaky blood vessels are formed which causes blindness in patient if untreated. Macugen binds to 165 isoform of VEGF (Vascular Endothelial Growth Factor) and stops its interactions with VEGF receptors present on blood vessels in eyes [Table/Fig-3]. The anti-angiogenesis effect of aptamer not only stops the excessive growth of blood vessels, but also prevents the formation of defective blood vessels which ultimately reduces swelling in eyes. Pegaptanib is given by intravitreal injection into the eyes, more specifically, into the vitreous humour part of eyes [16]. Poly lactic-co-glycolic acid (PLGA) microsphere is used to encapsulate the drug for its release [17].
Macugen for AMD treatment.
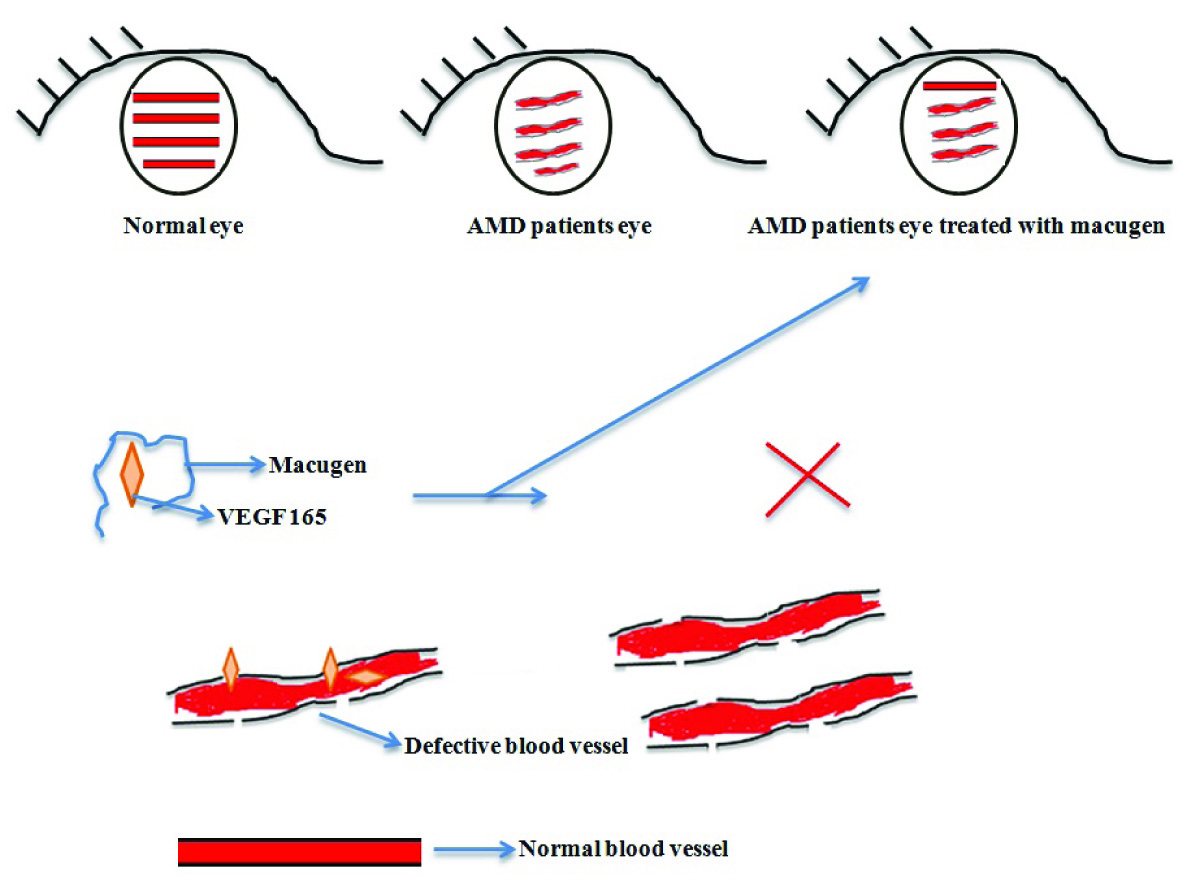
In animal studies it was also found that aptamer can be given subcutaneously and intravenously, while maintaining its desired concentration in blood [17]. In experimental studies macugen has shown inhibition of VEGF mediated vascular leakage which was measured by a corneal micropocket assay in guinea pigs (Eyetech Study Group, 2002). It has shown anti-angiogenesis effect in rat corneal angiogenesis model [18]. Further it has also effectively inhibited blood-retinal barrier breakdown in diabetic rat model [19]. In rhesus monkeys and rabbit models, macugen has shown neither the toxic effects nor change in intraocular pressure. Further, no immune response was shown by those animals [17,20]. In humans it has shown side effects such as anterior chamber inflammation, bleeding inside of the eyes, and infection of eyes or spots in vision. These issues can be minimized by adjusting the dose required by AMD affected person which vary from patient to patient. Pegaptanib has been approved in United States, Europe, Canada, Brazil and Australia [16,17,21]. Although this drug is used for AMD but a new monoclonal antibody (ranibizumab, Novartis) drug which is believed to be more effective than macugen is now available in market and competing with this aptamer.
ii) ARC1905- To fight against AMD, a new approach was developed by Ophthotech Company [22]. They used aptamer to target C5 protein which is involved in breakdown of membrane of cell. It has been known from past few years that AMD could be genetic in nature and caused by hyper action of alternate pathway of complement system. It is believed that mutation in chromosome 1q31 region expresses defective factor H protein of alternate complement system and is involved in causing AMD [23–28]. The over activation of complement system ultimately activates C5 protein that starts a cascade involving more complement proteins and forming Membrane Attack Complex (MAC) which ultimately kills retinal cells.
Clot Buster Aptamers
i) ARC183- It is a 15 bases long single stranded DNA aptamer [29] having G-quadruplex structure that interacts with exosite I of α-thrombin protein [30] [Table/Fig-4]. This interaction inhibits the binding of fibrinogen to thrombin causing anti-coagulation. The interaction is specific as it does not interact with other form of thrombin such as γ-thrombin. Further its half-life is only of two minutes creating rapid reversal of its own effect, thus it can be used in coronary artery bypass graft surgery. A more advanced version of this aptamer which is optimized and called as NU172 (26-mer DNA aptamer) is under phase II trial and soon be commercialized by ARCA biopharma Company [31].
Clot buster aptamers in use for cardiac operation.
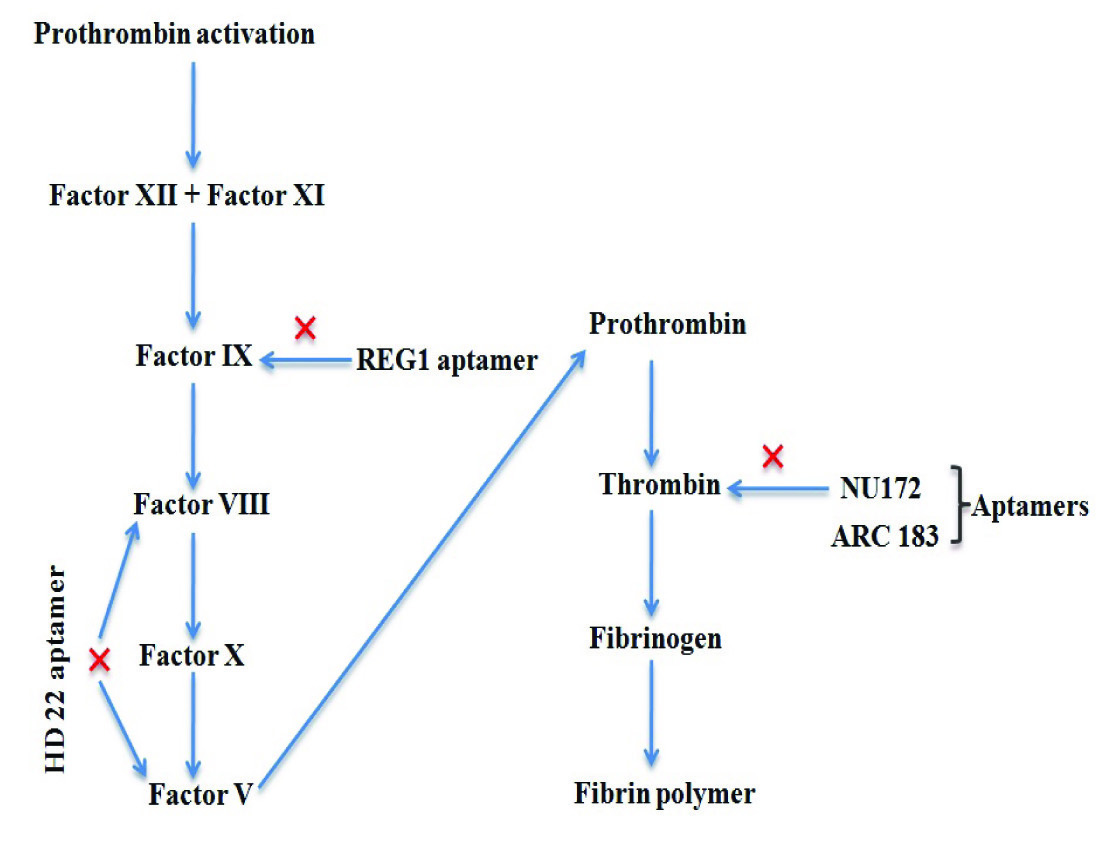
ii) Aptamer HD22- This aptamer contains duplex/G-quadruplex mixed structure which recognizes exosite II of thrombin and is involved in inactivation of factor V and factor VIII proteins of blood clotting system [32] [Table/Fig-4]. Further, the binding affinity of aptamer HD22 for thrombin is low as compared to ARC183.
iii) REG1 aptamer- This aptamer binds with factor IX and stops blood clotting process [Table/Fig-4]. This RNA aptamer is in trial process for its commercialization by Regado Biosciences Company [33].
iv) ARC1779 aptamer- It targets A1 domain of von Willebrand factor and is in phase II of clinical trial (Archemix Company) [34]. Nuclease resistant RNA aptamer selected against factor XII of intrinsic pathway could also be used in preventing pathological thrombosis formation [35]. One of the advantages of using these aptamers is that not only they can be used in therapeutics, but also as sensors for diagnosis of thrombosis by tagging fluorescent molecules with them.
Anti-obesity Aptamer
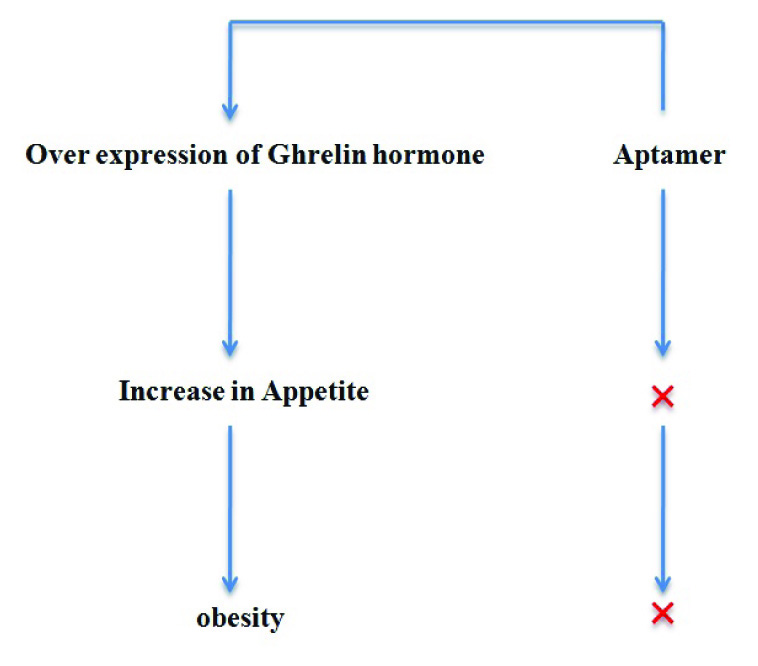
The NOXXON Company has developed spiegelmers (L-aptamer) against ghrelin, a peptide hormone associated with appetite and weight gain [Table/Fig-5]. This aptamer can serve as an anti-obesity drug and the animal test has already shown positive results in rats [36].
Spiegelmers against ghrelin hormone.
Aptamers Against Autoantibodies
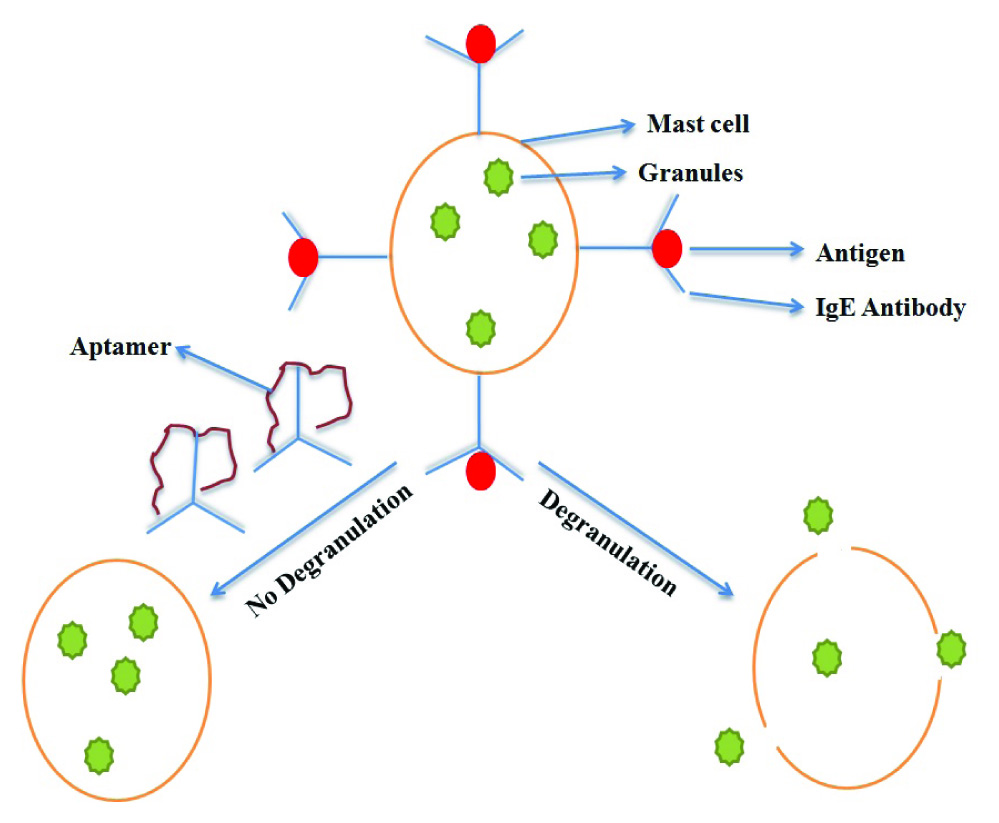
i) Role in allergic response- Immunoglobulin E (IgE) is involved in type 1 hypersensitivity reaction. Type 1 allergens can be due to environment factors (dust, grass, pollens) or food which activates immune system to produce IgE antibodies in excessive amounts. Overproduction of IgE causes asthma, allergies, dermatitis [37]. The molecular mechanisms involves allergens (antigen)-antibody interaction. Fc portion of IgE has receptors on mast cell. When immune complex (antigen+antibody) binds to mast cell, rupturing cell membrane releases large amounts of granules. These granules release mediators such as histamine, serotonin, prostaglandins, cytokines etc. which are involved in smooth muscle contraction, increase in vascular permeability or pulmonary smooth muscle contraction, ultimately causing hypertensive responses [Table/Fig-6]. Allergic responses can be stopped by inhibiting the interaction between antibody and mast cell. A DNA aptamer developed by Mendonsa group [38] in 2004 binds specifically to IgE antibody and masks its effect. Wang et al., developed aptamer sensor which is based on fluorescence protection assay for detection of IgE and the detection limit was 0.1 nM [39].
DNA aptamer binds specifically to IgE antibody.
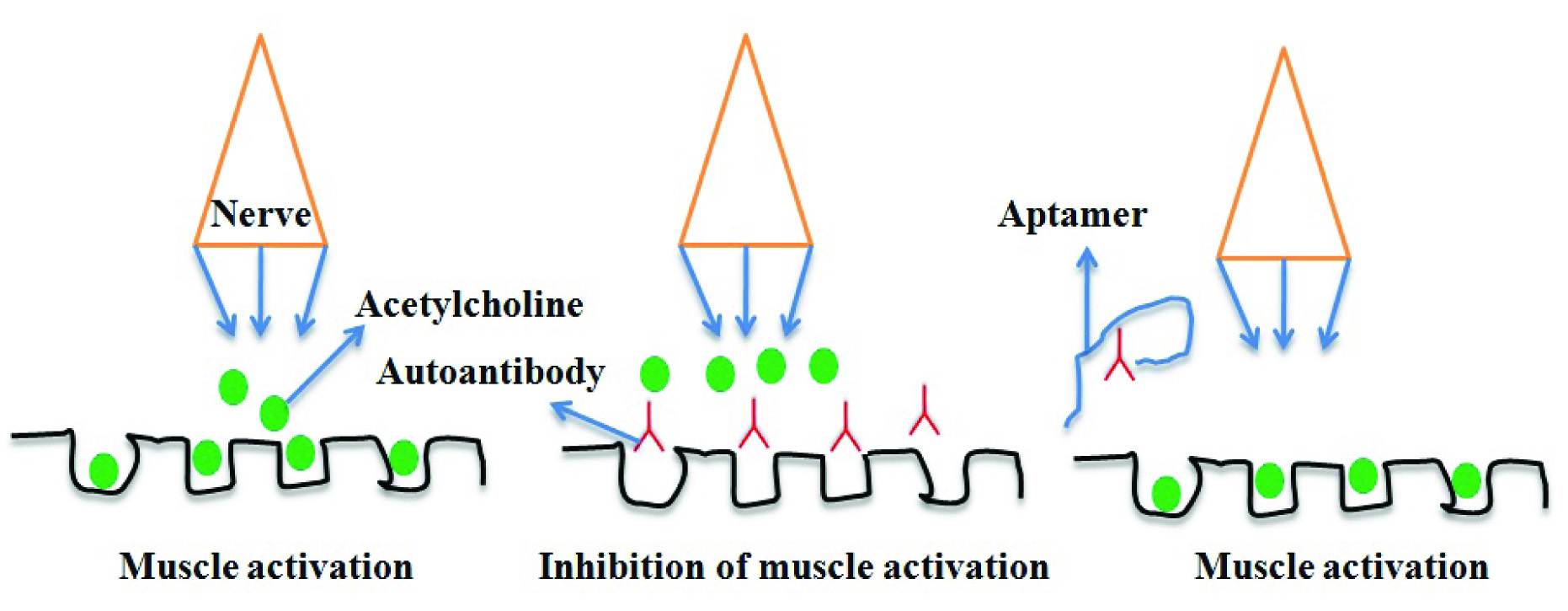
ii) Role in prevention of autoimmune disorders- Systemic Lupus Erythaematosus (SLE) is very common autoimmune disorder in women as compared to men. Auto-antibodies are generated against patient’s own DNA, RBC and platelet membrane. These antibodies cause lysis of RBC causing anaemia and activate complement system that are involved in inflammatory responses causing tissue damage. RNA aptamers have been selected against anti-DNA autoantibodies frequently found in SLE patients. The Kd value of aptamer is 2 nM and is highly specific to autoantibody [40]. In Myasthenia Gravis (MG), autoantibodies are generated against nicotinic acetylcholine esterase receptors (AChRs) found in skeletal muscle. The inhibition by autoantibody prevents the interaction between receptor and acetylcholine hormone, causing muscular weakness and fatigue. A RNA aptamer against AChRs autoantibodies has shown inhibition of autoimmune response in animal models of MG and also showed bioactive protection of AChRs in human cells [Table/Fig-7] [41]. Cho et al., has also selected an aptamer (RNA) specific to AChRs which can be useful for MG patient in near future [42].
DNA aptamer binds specifically to AChRs autoantibody.
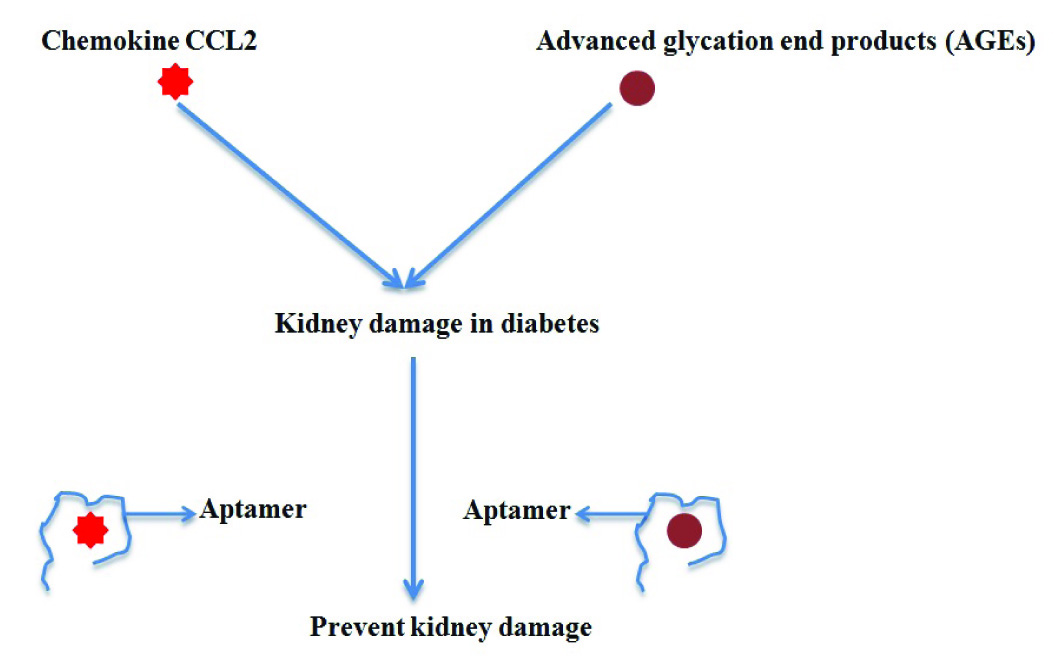
iii) Role in diabetes- Diabetes is life style disease and occurs most probably due to obesity, lack of physical activity; intake of high sugar or it could be genetic. Diabetes 1 is also known as childhood diabetes where patient fails to produce insulin due to destruction of beta cells of pancreas. In diabetes II (adult diabetes), insulin is produced properly but insulin receptors of cell become inactive due to presence of auto antibodies against them, causing inhibition of glucose metabolism. Lee and group selected a nuclease resistant aptamer (RNA sequence) that can bind to murine insulin receptor antibody (MA20). MA20 antibody destroys the insulin receptors of murine, causing diabetes [43]. The use of RNA aptamer has reduced the symptoms of diabetes in animal model and could be used for diabetes II treatment in humans, due to similarity between insulin receptor of murine and human. The main complication associated with diabetes II is damage of kidney which is also known as chronic kidney disease (CKD) which occurs due to action of chemokine CCL2 (MCP-1). NOX-E36 aptamer (RNA aptamer) targets CCL2 and reduces the progression of kidney damage [44–46] and is in phase II of clinical trial, developed by NOXXON pharma. In another work Japanese researcher raised and tested DNA aptamer against advanced glycation end products (AGEs) that cause nephropathy in diabetic patient. The experimental data is very promising and it is believed that in near future aptamer could be used for therapeutic purposes [Table/Fig-8] [47].
Aptamer to prevent diabetes nephropathy.
Role in Cancer Diagnosis and Therapy
Cancer is unregulated proliferation of cells which occurs due to gain in function or loss in function of genes. The cause of this change can be genetic or by environmental pollutants. Several methods are now available to fight against this deadly disease. [Table/Fig-9] depicts proteins that are overexpressed during cancer and selected aptamer against them that can be used for diagnosis or therapeutic purposes.
Aptamers against proteins involved in cancer.
| Target | Aptamer | Function of proteins in cancer |
|---|
| Pigpen | DNA | Endothelial protein, expresses on the surface of microvessels which is involved in blood vessel formation in cancer cells [48]. |
| PDGF-r | DNA | Platelet Derived Growth Factor Receptor (PDGF-r) is a tyrosine kinase protein involves in metastasis of tumour [49–51]. |
| Tenascin-C | DNA | This is an extracellular matrix protein that is expressed by chromosome 9 in humans. It is basically involved in cell signaling processes and activated during events such as fetal development, wound healing and in case of tumour growth [52–55]. |
| CTLA-4 | DNA | Cytotoxic T cell antigen-4 is a transmembrane protein that is expressed on the surface of activated T cells. Activated T cells further reduce the T cell response by raising threshold response needed for T-cell activation during cancer [56]. |
| Sialyl Lewis X | DNA | It is a tetrasaccharide carbohydrate that is attached to O-glycans on the surface of cells and selectively binds with selectin proteins during cell adhesion and inflammation. Sialyl Lewis X is overexpressed in cancer cells and through selectin helps in metastasis process of cancer [57]. |
| Nucleolin | DNA | It is a nucleolar phosphoprotein and found in nucleolus region of nucleus. Recent studies have shown the presence of this protein on cell surface especially on cancer cells. Thus nucleolin could be used as marker in cancer [58,59]. |
| PMSA | DNA | Prostate specific membrane antigen is a transmembrane protein and found at prostate epithelial cell and involved in hydrolytic cleavage of poly-γ-glutamated folates to produce glutamate. In general PMSA has high expression and activity in prostate tumour as compared to normal one which can be used as possible biomarker as well as target site for cancer treatment [60–64]. |
In hepatic neoplasm, Osteopontin (OPN) [65], Alfa-Feto Protein (AFP) and heterogeneous nuclear ribonucleoprotein A1 are involved in tumour growth and metastasis and could be used as target site for cancer treatment. Aptamers specific to these protein have shown drastic reduction in tumour formation in liver cell in in vitro conditions [66,67]. Xu and group in 2015 [68] selected seven aptamers through cell-SELEX method that are specific for liver cancer cell (HepG2 cell line) but not to normal cells. Further they also concluded that these aptamers also bind with lung cancer, ovarian cancer and luminal A subtype breast cancer cells, showing multi specific nature of these aptamer in recognizing cancer cells and could be used in cancer therapy. P12FR2 RNA aptamer is specific to pancreatic adenocarcinoma up-regulated factor (PAUF), and could be used in treatment of pancreatic cancer. This aptamer binds with PAUF and inhibit metastasis process in cancer [69]. Other aptamers such as NOX-A50 and NOX-f33 (polyethylene glycol-modified Spiegelmers) are specific to A1 High Mobility Group (HMGA1) proteins which are involved in anchorage-independent growth and epithelial-mesenchymal transition in normal cells making them cancerous one [70]. Epidermal growth factor receptor (EGFR) also found to be overexpressed in pancreatic and other types of cancer. Nuclease resistant RNA aptamer specific to EGFR was selected and it inhibits tumour growth in a mouse xenograft model of human non-small-cell lung cancer [71]. DNA aptamer, called XQ-2d was developed having high affinity and specificity for pancreatic ductal adenocarcinoma (PDAC). Aptamer XQ-2d selectively binds to PL45 cells and can be used in PDAC diagnosis and treatment [72].
Kwak et al., selected a RNA aptamer, specific to peroxisome proliferator-activated receptor delta (PPAR-delta) [73]. PPAR-delta is overexpressed in colon cancer which further helps in overexpression of vascular endothelial cell growth factor-A and cyclooxygenase-2. RNA aptamer has reduced tumourogenecity in HCT116 colon cancer cells by inhibiting expression of PPAR-delta protein. Another protein beta-catenin is involved in transcription and alternate splicing of oncogenic genes. High-affinity RNA aptamer has been developed against this protein which reduces the activation of these genes [74]. In one of the experiment Hungs et al., selected eight aptamers specific to colorectal cancer cells and stem cells by using on-chip cell-SELEX method [75].
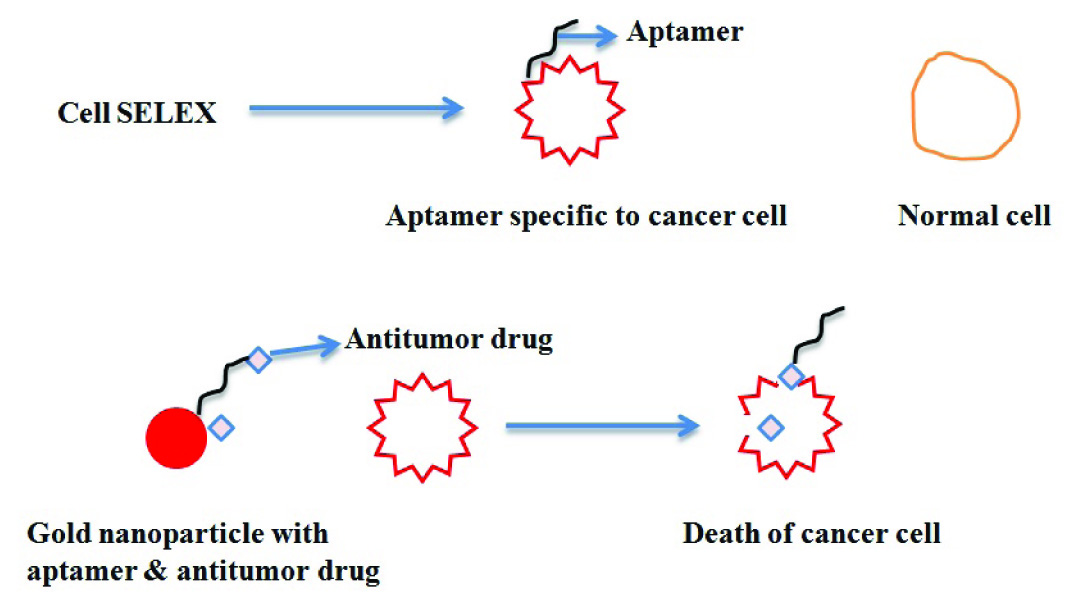
One of the interesting way of treating cancer is to select aptamer that are specific to cancer cells but not to normal cell through cell-SELEX. These selected aptamers can act as carrier for antitumour drug or toxin. Reports have shown that aptamer is first coated on carbon nanotubes or quantum dots or gold or iron nanoparticles and then therapeutic drug is either intercalated to aptamer or directly attached to nanoparticles [Table/Fig-10] [76].
Aptamer based cancer therapy.
Aptamers as Antiviral Agent
In past few years we have seen that viral infection is major cause of human death. New virus strains are emerging and at the same time antiviral drugs fail to respond, further side effects of these drugs have created need for search of new kind of drug. Aptamer could work in this direction. Viral infection can be inhibited by preventing virus fusion to human cell such as Hepatitis C virus which infects liver cells of human by interaction between E2 glycoprotein of virus and CD81 receptors of liver cell. A DNA aptamer was selected against E2 glycoprotein that inhibits the above interaction in in vitro studies [77]. Other approaches uses aptamers specific to viral polymerase enzymes [78], genetic material of virus [79], capsid proteins of virus [80] or other proteins that are involved in virus replication or processing of virus inside the cell. In one of the new approaches of cancer therapy aptamers are conjugated to small interfering (si) RNA to kill cancer T cell [56]. This approach can also be used to prevent viral infection.
Constrain of Using Aptamer in both in vitro and in vivo Conditions
Despite of its bright future in diagnostics and in therapeutics, aptamers are still in their preliminary stages of developments and there are constrains such as nuclease sensitivity, small size, toxic effect and transportation of aptamers to inside the cell which needs to be overcome before it can be used for in vitro and in vivo conditions.
i) Making nuclease resistance- There are many ways in which nuclease effects on aptamers can be reduced. Aptamers contain several sites such as sugar moiety, phosphodiester region where modification can be done without interrupting its activity, one example is capping of 3’ end of ssDNA as serum contains nucleases that act on 3’ end rather than on 5’ end. Substituting natural nucleotides with unnatural ones, such as 2’-F, 2’-OCH3 or 2’-NH2 modified nucleotides reduces affinity of nuclease for DNA degradation. Further, L-enantiomers form of nucleotides also known as spiegelmers makes aptamer nuclease resistant.
ii) Increasing renal filtration time- As already mentioned low size (10 to 15 kDa) causes quick filtration of aptamer from kidney, reducing its overall effect. Methods are being in used to increase the size of ssDNA by adding PEG (Poly Ethylene Glycol) or cholesterol. In case of macugen the PEGylation process increased the size from 10 kDa to 40kDa and at the same time half-life was also increased from 1 day to 10 days [81]. The increase in half time in serum shows that the renal filtration process was slow because of large size of modified aptamer.
iii) Reduction in toxicity- Very little information in this regard is available. As it is believed that aptamers are less immunogenic as compared to protein. But it’s difficult to predict that they are free from toxic effect, as already macugen has shown its side effects in AMD patient. It is believed that in near future when more and more aptamer based drugs will be used to treat patients then we will face the toxicity issue.
One of the disadvantages of aptamers is that it is difficult to select against those targets that are negatively charged and hydrophobic in nature as ssDNA molecules are also negatively charged. SELEX is the only technology which is being used for aptamers selection while there are well established methods available for antibody generation. Aptamer selection against small molecules is always a big challenge. In most cases those aptamers are selected that have affinity for target as well as immobilizer or linker attached to target, reducing aptamers selectivity. Also, the generation of aptamer is based on combinatorial DNA library which contains random sequences of aptamers but it seems difficult to match diversity that is found in immune system of living beings used for antibody generation.
Conclusion
Aptamers are short oligonucleotides of DNA or RNA selected in vitro to bind a specific target with high specificity and high affinity which require simple synthesis protocols compared to the in vivo development of antibodies. It might become an alternative to monoclonal antibodies due to its flexibility of in vitro selection. Yet, despite the advances and the huge body of literature documenting the success of the technology, the commercial application of aptamers remains relatively undeveloped. The fact that there is a vast antibody-based market and a certain degree of hesitation to move to a new type of product, unless aptamers offer verifiably significant improvements on current technologies. In recent years advances are being made to improve the efficiency of selection and to increase the affinity, specificity, biostability and bioavailability of aptamers.
Aptamers are emerging as promising bio-recognition elements for diagnostic, therapeutic and biosensing purposes. It is likely that in near future aptamer technology will increasingly find use in the development of new therapeutic and diagnostic agents.
[1]. Tuerk C, Gold L, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymeraseSci 1990 3:505-10. [Google Scholar]
[2]. Song KM, Lee S, Ban C, Aptamers and their biological applicationsSensors 2012 12:612-31. [Google Scholar]
[3]. Sundaram P, Kurniawan H, Byrne ME, Wower J, Therapeutic RNA aptamers in clinical trialsEur J Pharm Sci 2013 48:259-71. [Google Scholar]
[4]. Ellington AD, Szostak JW, Invitro selection of RNA molecules that bind specific ligandsNat 1990 346(6287):818-22. [Google Scholar]
[5]. Parashar A, Rajput YS, Sharma R, Aptamer-based sensing of betacasomorphin-7J Agric Food Chem 2015 63(10):2647-53. [Google Scholar]
[6]. Hünniger T, Fischer C, Wessels H, Hoffmann A, Kratzin AP, Haase I, Food sensing: selection and characterization of DNA aptamers to Alicyclobacillus spores for trapping and detection from orange juiceJ Agric Food Chem 2015 63(8):2189-97. [Google Scholar]
[7]. Mehta J, Martin ER, Dorst BV, Maes B, Herrebout W, Scippo ML, Selection and characterization of PCB-binding DNA aptamersAnal Chem 2012 84(3):1669-76. [Google Scholar]
[8]. Parekh P, Tang Z, Turner PC, Moyer RW, Tan W, Aptamers recognizing glycosylated haemagglutinin expressed on the surface of vaccinia virus-infected cellsAnal Chem 2010 82(20):8642-49. [Google Scholar]
[9]. Luo Y, He L, Zhan S, Wu Y, Liu L, Zhi W, Ultrasensitive resonance scattering (RS) spectral detection for trace tetracycline in milk using aptamer-coated nanogold (ACNG) as a catalystJ Agric Food Chem 2014 62:1032-37. [Google Scholar]
[10]. Liu J, Liu H, Sefah K, Liu B, Pu Y, Van Simaeys DV, Selection of aptamers specific for adipose tissuePLoS ONE 2012 7(5):e37789 [Google Scholar]
[11]. Malhotra S, Pandey AK, Rajput YS, Sharma R, Selection of aptamers for aflatoxin M1 and their characterizationJ Mol Recognit 2014 27:493-500. [Google Scholar]
[12]. Chang EK, Eckert MA, Ali MM, Riazifar H, Pone EJ, Liu L, Facile supermolecular aptamer inhibitors of L-SelectinPLoS ONE 2015 10(3):e0123034 [Google Scholar]
[13]. Ramos P, Soto A, Martin S, A DNA aptamer population specifically detects Leishmania H2A antigenLab Invest 2007 87:409-16. [Google Scholar]
[14]. Ng EWM, Shima DT, Calias P, Cunningham ET, Guyer DR, Adamis AP, Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular diseaseNature Reviews Drug Discovery 2006 5:123-32. [Google Scholar]
[15]. Kourlas H, Schiller DS, Pegaptanib sodium for the treatment of neovascular age-related macular degeneration: A ReviewClinical Ther 2006 28:36-44. [Google Scholar]
[16]. “Macugen (pegaptanib)” (PDF)European Medicines Agency 2010 :1-3. [Google Scholar]
[17]. Vinores SA, Pegaptanib in the treatment of wet, age-related macular degenerationInt J Nanomedicine 2006 1(3):263-68. [Google Scholar]
[18]. Proske D, Blank M, Buhmann R, Resch A, Aptamers -basic research, drug development, and clinical applicationsAppl Microbiol Biotechnol 2005 69:367-74. [Google Scholar]
[19]. Ishida S, Usui T, Yamashiro K, Kaji Y, Amano S, Ogura Y, VEGF164-mediated inflammation is required for pathological, but not physiological, ischemia–induced retinal neovascularizationJ Exp Med 2003 198:483-89. [Google Scholar]
[20]. Vinores SA, Technology evaluation: pegaptanib, Eyetech/PfizerCurr Opin Mol Ther 2003 5:673-79. [Google Scholar]
[21]. “Eyetech announces approval of macugen(R) in Brazil for the treatment of neovascular (wet) age-related macular degeneration”Evaluate 2005 [Google Scholar]
[22]. Biesecker G, Dihel L, Enney K, Bendele RA, Derivation of RNA aptamer inhibitors of human complement C5Immunopharmacology 1999 42(1-3):219-30. [Google Scholar]
[23]. Rodriguez de Cordoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P, The human complement factor H: functional roles, genetic variations and disease associationsMol Immunol 2004 41(4):355-67. [Google Scholar]
[24]. Souied EH, Leveziel N, Richard F, Dragon-Durey MA, Coscas G, Soubrane G, Y402H complement factor H polymorphism associated with exudative age-related macular degeneration in the French populationMol Vis 2005 11:1135-40. [Google Scholar]
[25]. Okamoto H, Umeda S, Obazawa M, Minami M, Noda T, Mizota A, Complement factor H polymorphisms in Japanese population with age-related macular degenerationMol Vis 2006 12:156-58. [Google Scholar]
[26]. Seitsonen S, Lemmela S, Holopainen J, Tommila P, Ranta P, Kotamies A, Analysis of variants in the complement factor H, the elongation of very long chain fatty acids-like 4 and the hemicentin 1 genes of age-related macular degeneration in the Finnish populationMol Vis 2006 12:796-801. [Google Scholar]
[27]. Simonelli F, Frisso G, Testa F, di Fiore R, Vitale DF, Manitto MP, Polymorphism p. 402Y > H in the complement factor H protein is a risk factor for age related macular degeneration in an Italian populationBr J Ophthalmol 2006 90(9):1142-45. [Google Scholar]
[28]. Li M, Atmaca-Sonmez P, Othman M, Branham KE, Khanna R, Wade MS, CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degenerationNat Genet 2006 38(9):1049-54. [Google Scholar]
[29]. Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ, Selection of single-stranded DNA molecules that bind and inhibit human thrombinNat 1992 35:564-66. [Google Scholar]
[30]. Paborsky LR, McCurdy SN, Griffin LC, Toole JJ, Leung LL, The single-stranded DNA aptamer-binding site of human thrombinJ Biol Chem 1993 268:20808-11. [Google Scholar]
[31]. Waters EK, Richardson J, Schaub RG, Kurz JC, Effect of NU172 and bivalirudin on ecarin clotting time in human plasma and whole bloodJ Thromb Haemost 2009 (Supplement 2)Abstract PPWE-168 [Google Scholar]
[32]. Tasset DM, Kubik MF, Steiner W, Oligonucleotide inhibitors of human thrombin that bind distinct epitopesJ Mol Biol 1997 272(5):688-98. [Google Scholar]
[33]. Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, RNA aptamers as reversible antagonists of coagulation factor IXaNat 2002 419(6902):90-94. [Google Scholar]
[34]. Gilbert JC, DeFeo-Fraulini T, Hutabarat RM, Horvath CJ, Merlino PG, Marsh HN, First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteersCirculation 2007 116(23):2678-86. [Google Scholar]
[35]. Woodruff RS, Xu Y, Layzer J, Wu W, Ogletree ML, Sullenger BA, Inhibiting the intrinsic pathway of coagulation with a factor XII–targeting RNA aptamerJ Thromb Haemost 2013 11:1364-73. [Google Scholar]
[36]. Helmling S, Christian M, Eulberg D, Buchner K, Schröder W, Lange C, Inhibition of ghrelin action invitro and invivo by an RNA-SpiegelmerPNAS 2004 101(36):13174-79. [Google Scholar]
[37]. Gould HJ, Sutton BJ, Beavil AJ, Beavil RL, McCloskey N, Coker HA, The biology of IGE and the basis of allergic diseaseAnnu Rev Immunol 2003 21:579-28. [Google Scholar]
[38]. Mendonsa SD, Bowser MT, Invitro Evolution of functional DNA using capillary electrophoresisJ Am Chem Soc 2004 126(1):20-21. [Google Scholar]
[39]. Wang HQ, Wu Z, Tang LJ, Yu RQ, Jiang JH, Fluorescence protection assay: a novel homogeneous assay platform toward development of aptamer sensors for protein detectionNucleic Acids Res 2011 39(18):e122 [Google Scholar]
[40]. Kim YM, Choi KH, Jang YJ, Yu J, Jeong S, Specific modulation of the anti-DNA autoantibody-nucleic acids interaction by the high affinity RNA aptamerBiochem Biophys Res Commun 2003 300(2):516-23. [Google Scholar]
[41]. Hwang B, Lee SW, Improvement of RNA aptamer activity against myasthenic autoantibodies by extended sequence selectionBiochem Biophys Res Commun 2002 290(2):656-62. [Google Scholar]
[42]. Cho JS, Lee SW, Sequence and structural features of RNA aptamer against myasthenic autoantibodiesOligonucleotides 2009 19(3):273-80. [Google Scholar]
[43]. Lee SW, Sullenger BA, Isolation of a nuclease–resistant decoy RNA that selectively blocks autoantibody binding to insulin receptors on human lymphocytesJ Exp Med 1996 184:315-24. [Google Scholar]
[44]. Oberthür D, Achenbach J, Gabdulkhakov A, Buchner K, Maasch C, Falke S, Crystal structure of a mirror-image L-RNA aptamer (Spiegelmer) in complex with the natural L-protein target CCL2Nat Commun 2015 6:6923 [Google Scholar]
[45]. Ninichuk V, Clauss S, Kulkarni O, Schmid H, Segerer S, Radomska E, Late onset of Ccl2 blockade with the spiegelmer mNOXE36- 3′PEG prevents glomerulosclerosis and improves glomerular filtration rate in db/db miceAm J Pathol 2008 172(3):628-37. [Google Scholar]
[46]. Maasch C, Buchner K, Eulberg D, Vonhoff S, Klussmann S, Physicochemical stability of NOX-E36, a 40mer L-RNA (Spiegelmer) for therapeutic applicationsNucleic Acids Symp Ser (Oxf) 2008 (52):61-62. [Google Scholar]
[47]. Kaida Y, Fukami K, Matsui T, Higashimoto Y, Nishino Y, Obara N, DNA aptamer raised against AGEs blocks the progression of experimental diabetic nephropathyDiabetes 2013 62:3241-50. [Google Scholar]
[48]. Blank M, Weinschenk T, Priemer M, Schluesener H, Systematic evolution of a DNA aptamer binding to rat brain tumour microvessels selective targeting of endothelial regulatory protein pigpenJ Biol Chem 2001 276:16464-68. [Google Scholar]
[49]. Pietras K, Rubin K, Sjoblom T, Buchdunger E, Sjoquist M, Heldin CH, Inhibition of platelet-derived growth factor receptors reduces interstitial hypertension and increases transcapillary transport in tumoursCancer Res 2001 61:2929-34. [Google Scholar]
[50]. Pietras K, Rubin K, Sjoblom T, Buchdunger E, Sjoquist M, Heldin CH, Inhibition of PDGF receptor signaling in tumour stroma enhances antitumour effect of chemotherapyCancer Res 2002 62:5476-84. [Google Scholar]
[51]. Green LS, Jellinek D, Jenison R, Ostman A, Heldin CH, Janjic N, Inhibitory DNA ligands to platelet derived growth factor B-chainBiochemistry 1996 35(45):14413-24. [Google Scholar]
[52]. Herold-Mende C, Mueller MM, Bonsanto MM, Schmitt HP, Kunze S, Steiner HH, Clinical impact and functional aspects of tenascin-C expression during glioma progressionInt J Cancer 2002 98(3):362-69. [Google Scholar]
[53]. Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, Tenascin-C aptamers are generated using tumour cells and purified proteinJ Biol Chem 2001 276:48644-54. [Google Scholar]
[54]. Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, Application of locked nucleic acids to improve aptamer invivo stability and targeting functionNucleic Acids Res 2004 32:5757-65. [Google Scholar]
[55]. Dion DA, Chen H, Hicke BJ, Swiderek KM, Gold L, A tenascin-C aptamer identified by tumour cell SELEX: Systematic evolution of ligands by exponential enrichmentPNAS 2003 100(26):15416-21. [Google Scholar]
[56]. Herrmann A, Priceman SJ, Swiderski P, Kujawski M, Xin H, Cherryholmes GA, CTLA4 aptamer delivers STAT3 siRNA to tumour-associated and malignant T cellsJ Clin Invest 2014 124(7):2977-87. [Google Scholar]
[57]. Jeong S, Eom T, Kim S, Lee S, Yu J, Invitro selection of the RNA aptamer against the Sialyl Lewis X and its inhibition of the cell adhesionBiochem Biophys Res Commun 2001 281:237-43. [Google Scholar]
[58]. Christian S, Pilch J, Akerman ME, Porkka K, Laakkonen P, Ruoslahti E, Nucleolin expressed at the cell surface is a marker of endothelial cells in angiogenic blood vesselsJ Cell Biol 2003 163:871-78. [Google Scholar]
[59]. Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO, Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancerExp Mol Pathol 2009 86(3):151-64. [Google Scholar]
[60]. Carter RE, Feldman AR, Coyle JT, Prostate-specific membrane antigen is a hydrolase with substrate and pharmacologic characteristics of a neuropeptidaseProc Natl Acad Sci USA 1996 93(2):749-53. [Google Scholar]
[61]. Pinto JT, Suffoletto BP, Berzin TM, Qiao CH, Lin S, Tong WP, Prostate-specific membrane antigen: a novel folate hydrolase in human prostatic carcinoma cellsClin Cancer Res 1996 2(9):1445-51. [Google Scholar]
[62]. Lapidus RG, Tiffany CW, Isaacs JT, Slusher BS, Prostate-specific membrane antigen (PSMA) enzyme activity is elevated in prostate cancer cellsProstate 2000 45(4):350-54. [Google Scholar]
[63]. Burger MJ, Tebay MA, Keith PA, Samaratunga HM, Clements J, Lavin MF, Expression analysis of delta-catenin and prostate-specific membrane antigen: their potential as diagnostic markers for prostate cancerInt J Cancer 2002 100(2):228-37. [Google Scholar]
[64]. Lupold SE, Hicke BJ, Lin Y, Coffey DS, Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigenCancer Res 2002 62:4029-33. [Google Scholar]
[65]. Bhattacharya SD, Mi Z, Kim VM, Guo H, Talbot LJ, Kuo PC, Osteopontin regulates epithelial mesenchymal transition-associated growth of hepatocellular cancer in a mouse xenograft modelAnn Surg 2012 255(2):319-25. [Google Scholar]
[66]. Lee YJ, Lee SW, Regression of hepatocarcinoma cells using RNA aptamer specific to alpha-fetoproteinBiochem Biophys Res Commun 2012 417:521-27. [Google Scholar]
[67]. Li S, Wang W, Ding H, Xu H, Zhao Q, Li J, Aptamer BC15 against heterogeneous nuclear ribonucleoprotein A1 has potential value in diagnosis and therapy of hepatocarcinomaNucleic Acid Ther 2012 22(6):391-98. [Google Scholar]
[68]. Xu J, Teng IT, Zhang L, Delgado S, Champanhac C, Cansiz S, Molecular recognition of human liver cancer cells using DNA aptamers generated via cell-SELEXPLoS ONE 2015 10(5):e0125863 [Google Scholar]
[69]. Kim YH, Sung HJ, Kim S, Kim EO, Lee JW, Moon JY, An RNA aptamer that specifically binds pancreatic adenocarcinoma up-regulated factor inhibits migration and growth of pancreatic cancer cellsCancer Lett 2011 313(1):76-83. [Google Scholar]
[70]. Maasch C, Vater A, Buchner K, Purschke WG, Eulberg D, Vonhoff S, Polyetheylenimine-polyplexes of Spiegelmer NOX-A50 directed against intracellular high mobility group protein A1 (HMGA1) reduce tumour growth invivoJ Biol Chem 2010 285(51):40012-18. [Google Scholar]
[71]. Esposito CL, Passaro D, Longobardo I, Condorelli G, Marotta P, Affuso A, A neutralizing RNA aptamer against EGFR causes selective apoptotic cell deathPLoS ONE 2011 6(9):e24071 [Google Scholar]
[72]. Wu X, Zhao Z, Bai H, Fu T, Yang C, Hu X, DNA aptamer selected against pancreatic ductal adenocarcinoma for invivo imaging and clinical tissue recognitionTheranostics 2015 5(9):985-94. [Google Scholar]
[73]. Kwak H, Hwang I, Kim JH, Kim MY, Yang JS, Jeong S, Modulation of transcription by the peroxisome proliferator-activated receptor delta—binding RNA aptamer in colon cancer cellsMol Cancer Ther 2009 8(9):2664-73. [Google Scholar]
[74]. Lee HK, Choi YS, Park YA, Jeong S, Modulation of oncogenic transcription and alternative splicing by beta-catenin and an RNA aptamer in colon cancer cellsCancer Res 2006 66:10560-66. [Google Scholar]
[75]. Hung LY, Wang CH, Che YJ, Fu CY, Chang HY, Wang K, Screening of aptamers specific to colorectal cancer cells and stem cells by utilizing on-chip cell-SELEXNature (Scientific Reports) 2015 5:10326 [Google Scholar]
[76]. Zhou J, Rossi JJ, Cell-type-specific, aptamer-functionalized agents for targeted disease therapyMolecular Therapy-Nucleic Acids 2014 3:e169 [Google Scholar]
[77]. Yang D, Meng X, Yu Q, Xu L, Long Y, Liu B, Inhibition of Hepatitis C virus infection by DNA aptamer against envelope proteinAntimicrob Agents Chemother 2013 57(10):4937-44. [Google Scholar]
[78]. Ellingham M, Bunka DHJ, Rowlands DJ, Stonehouse NJ, Selection and characterization of RNA aptamers to the RNA-dependent RNA polymerase from foot-and-mouth disease virusRNA 2006 12(11):1970-79. [Google Scholar]
[79]. Konno K, Fujita S, Iizuka M, Nishikawa S, Hasegawa T, Fukuda K, Isolation and characterization of RNA aptamers specific for the HCV minus-IRES domain INucleic Acids Symp Ser 2008 52:493-94. [Google Scholar]
[80]. Orabi A, Bieringer M, Geerlof A, Bruss V, An Aptamer against the matrix binding domain on the Hepatitis B virus capsid impairs virion formationJ Virol 2015 89(18):9281-87. [Google Scholar]
[81]. Keefe AD, Pai S, Ellington A, Aptamers as therapeuticsNat Rev Drug Discov 2010 9(7):537-50. [Google Scholar]