Urinary Tract Infections (UTIs) are the most prevalent bacterial infections in India and worldwide. Escherichia coli are the most common bacteria present in the UTIs. The bacterium causes diseases in intestinal and extra-intestinal environments through acquired virulence factors through horizontal gene transfer, genetic recombination and natural selection [1,2] Uropathogenic E. coli (UPEC), which colonizes the urinary tract, may ascend the ureters to the kidney and establishes a secondary infection, acute pyelonephritis with irreversible kidney damage and causes of community acquired and nosocomial infections both in children and adults [3]. The wide spread multidrug resistant ESBL producing E. coli among UTIs gave an eye opening all over the world particularly in India [4–7].
The susceptibility of uropathogens to various antibiotics or its antibiogram profiling may help to improve the treatment of UTI without any delay. However there are many microorganisms responsible for UTI, E. coli is frequently present at community level infection. Among this, the persistent of high rate of resistant ESBL species gained much attention. The practice of irrational usage of antibiotics is one of the reasons to exhibit unique microorganism resistant pattern. Only few reports are available for the prevalence of multidrug resistant strains of UTI E. coli in the state of Tamil Nadu [8].
Phylogenetic studies based on multilocus enzyme electrophoresis (MLEE) showed that E. coli strains can be assigned to one of the four major phylogenetic groups (A, B1, B2 and D), which was later determined by PCR based method [9]. These four major groups become dominant recent years [10]. Among the four, extra intestinal pathogenic E. coli, which includes UPEC, comes under the group B2 and some extent to group D and the other two groups predominantly present as commensal and/or intestinal pathogens. The ancestral backbone genome that remains constant, whereas the genes which differentiate the pathogenicity were introduced in the later recent years of evolution [11]. The genetic analysis using RAPD typing is one of the better tools for analysing the genetic correlation among the closely related bacterial population [12]. RAPD was reported more sensitive than PFGE, southern blotting with insertion sequence probes and phage typing with respect to MLEE for the study of bacterial population genetic structure, evolution and epidemiology [13,14]. The antibiotic sensitivity and molecular typing will give a pathway for the treatment of UTIs.
The present work was aimed to evaluate the persistence of multiple drug resistant strains of E. coli isolated from UTIs in Tamil Nadu, irrespective of age and sex through morphological, biochemical and antibiogram analysis. In addition to that we examined the genetic correlation of phylogenetic UPEC isolates using RAPD analysis to understand the molecular typing.
Materials and Methods
Isolation and Identification of E. coli
During the period of 2011-2012, nearly 300 midstream urine samples were collected from the outpatients having symptoms of UTIs such as fever with irritation and pain during urination and a total number of 58 non-duplicate E. coli strains were isolated and identified biochemically. All the cultures were maintained and stored as glycerol stock culture at -80°C. Ethical clearance was obtained from Institutional Committee of Madurai Kamaraj University Ethical Committee for the collection of urine samples.
Antibiotic susceptibility test
All E. coli isolates were checked for their susceptibility towards different antibiotics on Muller-Hinton agar (Himedia, Mumbai), by disk diffusion method based on Clinical Laboratory Standard Institute (CLSI) guidelines [15], using E. coli ATCC 25922 as standard. The list of antibiotic discs used were ampicillin (AMP) 10μg, amoxycillin/ clavulanic acid (AMC) 20/10μg, ceftazidime (CAZ) 30μg, cefepime (CPM) 30μg, cefotaxime (CTX) 30μg, cefoxitin (CX) 30μg, ceftriaxone (CTR) 30μg, cefixime (CFM) 5μg, imipenem (IPM) 10μg, meropenem (MRP) 10μg, gentamycin (GEN) 10μg, nalidixic acid (NAL) 30μg, ciprofloxacin (CIP) 5μg, Co-trimaxazole (COT) 1.25/23.75μg, tetracycline (TE) 30μg, and nitrofurantoin (NIT) 300μg (HiMedia, Mumbai).
Phenotypic confirmation of ESBL production
Extended-Spectrum Beta-Lactamase (ESBL) was detected by the double-disk synergy (DDS) test using ceftazidime (30μg) and ceftazidime/clavulanic acid (30/10μg) as recommended by the CLSI [15]. An isolate was graded as ESBL producer when its zone of inhibition varied by ≥5mm with clavulanic acid. E. coli ATCC 35218 and E. coli ATCC 25922 were used as positive and negative controls respectively.
Phenotypic detection of AmpC β-lactamase
Cefoxitin resistant strains were analysed by combined disc method using the combination of cefoxitin (30μg) and cefoxitin/phenyl boronic acid (30/400μg), for the production of AmpC β-lactamase. ≥5mm increase in zone diameter with the presence of phenyl boronic acid against cefoxitin alone was considered positive for the production of AmpC β-lactamase. E. coli ATCC 25922 was used as a negative control [16]
Phylogenetic grouping and identification of ST131
The isolates were classified into any one the four major phylogenetic groups (A, B1, B2 and D) by multiplex PCR as done previously [9]. Phylogenetic group B2 strains were screened further for the presence of pabB genes to identify O25b-ST131 strains [17].
Random Amplified Polymorphic DNA (RAPD) Analysis
RAPD analysis was carried out for phylogenetic group B2 isolates with primer 1281 (5’-AACGCGCAAC-3’) to analyse the intra-species variation [18]. The dendrogram was constructed with PyElph 1.4 software [19].
Results
Prevalence of E. coli among UTI patients of Southern India
Nearly 58 E. coli strains were isolated and confirmed from the patients of different age, having the symptoms of UTI in Tamil Nadu, India during the period of December 2011 to June 2012. It is noteworthy that 24 strains were isolated from male and 34 from female patients. [Table/Fig-1] represented the prevalence of E. coli among different age and sex. The incidence of UTI was higher among female compared to male patients as prevalence was increased the age increased in male. Among female belonging to age group of 11-30 years were more susceptible to E. coli in UTIs with 24.1% followed by 31 to 50 years with 17.2% above 51 years with 10.3% and ≥50 years with 9%. Among male patients the incidence of UTI was higher among the elder people. A 19% of the isolates were from the age group of above 50 years, followed by 9% from 31 to 50 years, 7% from below 10 years and 3% from 11 to 30 years.
The Demographic profile of Antibiotic resistance of E. coli strains isolated from Urinary tract infection (UTI) patients.(Cip- Ciprofloxacin, Cot-Cotrimaxazole, Ctr-Ceftriaxone).
| E. coli Strains | Age (years) | Sex | Antibiotic Resistance | Beta lactamase |
|---|
| Cip | Cot | Ctr | ESBL | AmpC |
|---|
| XA01 | 71 | M | + | + | + | + | - |
| XA03 | 32 | F | + | + | + | + | + |
| XA04 | 57 | M | + | + | + | + | - |
| XA05 | 41 | F | + | + | + | + | + |
| XA06 | 25 | F | + | + | + | + | - |
| XA08 | 23 | M | + | + | - | + | + |
| XA09 | 38 | F | - | + | + | + | - |
| XA11 | 36 | F | + | + | + | + | - |
| XA12 | 22 | F | + | + | + | + | - |
| XA13 | 56 | M | + | + | + | + | - |
| XA19 | 0.7 | M | + | + | - | + | - |
| XA21 | 26 | F | + | + | + | + | - |
| XA24 | 45 | F | + | - | + | + | - |
| XA27 | 34 | F | + | + | + | + | - |
| XA31 | 17 | F | + | + | + | + | + |
| XA33 | 2 | M | + | + | + | + | - |
| XA34 | 52 | F | - | + | + | + | - |
| XA36 | 0.4 | F | + | - | + | + | - |
| XA37 | 0.9 | M | + | + | + | + | - |
| XA40 | 50 | M | + | + | + | + | - |
| XA42 | 64 | M | + | + | + | + | - |
| XA43 | 28 | F | + | + | + | + | - |
| XA46 | 62 | F | + | - | + | + | - |
| XA50 | 9 | M | + | - | + | + | - |
| XA51 | 56 | M | + | + | + | + | + |
| XA55 | 7 | F | + | + | + | + | + |
| XA58 | 34 | F | + | - | + | + | + |
Antibiotic Sensitivity profile
The sensitivity of the UPEC isolates towards 16 different antibiotics of different classes were analysed by disc diffusion method. All the isolates were sensitive towards metallo-β-lactamase antibiotics such as imipenem and meropenem, whereas none of them to amoxicillin/clavulanic acid. Most of the isolates were sensitive to nitrofurantoin (77.58%) followed by cefoxitin (65.51%), gentamycin (55.17%), ceftriaxone (48.27%), ceftazidime (46.55%), cefepime (41.37%), co-trimaxazole (34.48%), ciprofloxacin (24.13%), ampicillin (20.69%), cefixime (13.8%) and ceftriazone (12.07%) and tetracycline (1.72%) The antibiotic sensitivity profile is shown in [Table/Fig-2].
Antibiogram profiling of ESBL and non-ESBL isolates towards various antibiotics (from inner circle) AMP, AMC, CX, CTR, CAZ, CFM, CTX, CPM, IMP, MER, CIP, COT, GE, NA, NIT and TE.
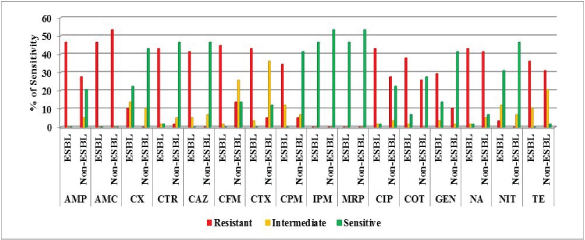
Resistance against quinolones was found higher with 84.48% for nalidixic acid and 70.69% for ciprofloxacin. Resistance to other group of antibiotics was also recorded higher with ampicillin (74.13%), tetracycline (67.24%), trimethoprim/ sulphamethoxazole (63.79%) and gentamycin (39.66%). Among the third generation cephalosporins, cefixime showed (58.62%) highest resistance, followed by cefotaxime (48.27%), ceftriaxone (44.82%) and ceftazidime (41.38%). When compared with third generation cephalosporins fourth generation cefepime was less resistance (39.66%).
Phenotypic detection of ESBL and AmpC β-lactamase production
Phenotypic detection of ESBL was performed for 28 cephalosporin resistant isolates out of which 27 (47%) isolates were identified to produce ESBL based on combined disc method. All ESBL isolates were resistant to ampicillin and cephalosporins except one isolate, E. coli XA19 which was sensitive for ceftriaxone. Along with cephalosporins, co-resistance to other groups such as quinolones (43.1% each for nalidixic acid and ciprofloxacin), sulphonamides (37.93 %), tetracycline (36.2%) and gentamycin (29.31%) was also higher among the ESBL isolates. All ESBL isolates were sensitive towards metallo-β-lactams. Among ESBL isolates, higher amount of sensitivity was found for nitrofurantoin and cefoxitin (31 and 24% respectively).
Cefoxitin resistance was directly correlated to ampC β-lactamase production. So isolates resistant (n=7) or intermediate resistant (n=11) to cefoxitin were tested for the production of AmpC β-lactamase using phenyl boronic acid as enzyme inhibitor. All 7 cefoxitin resistant isolates (E. coli XA03, E. coli XA05, E. coli XA08, E. coli XA55, E. coli XA31, E. coli XA51 and E. coli XA58) were identified to produce AmpC β-lactamase. Among these co-production of ESBL was detected in 6 isolates with the exception of XA08. All AmpC producing isolates were also resistant to other antibiotics such as extended spectrum cephalosporins, quinolones, tetracycline and aminoglycosides except one isolate (XA08), which showed intermediate resistance against cephalosporins.
Phylogenetic grouping and molecular typing
According to Clermont et al., the PCR based phylogenetic analysis was carried out for all UTI E. coli isolates [9]. Among these 24 isolates (41%) were assigned as group B2, 19 isolates (33%) as groups A, 8 isolates (14%) as B1 and 7 isolates (12%) as group D.
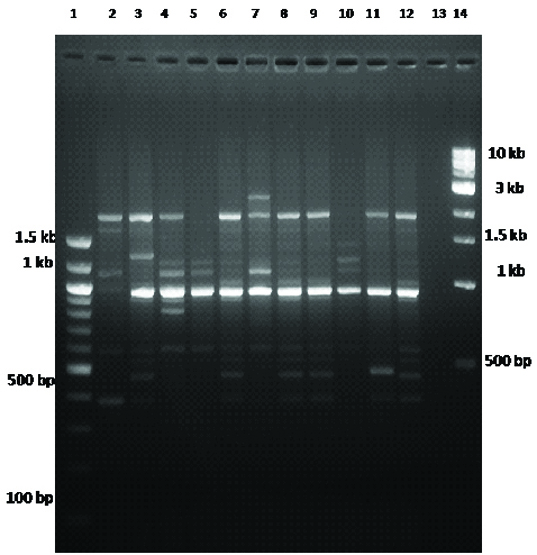
PCR based molecular typing was carried out based on RAPD analysis using primer 1281 for randomly selected isolates [Table/Fig-3]. All isolates from group B2 were included in this analysis along with 4, 6 and 3 isolates from group A, B1 and D respectively. The amplified product ranged from 400 bp to 3 kb with upto 12 bands. The dendrogram was constructed based on unweighed pair group method of analysis (UPGMA), which revealed the genetic correlation among isolates [Table/Fig-4]
RAPD analysis of randomly selected UTI E. coli strains using primer 1281. Lane1 and 14 are molecular makers. Lane2-12 are randomly selected E. coli strains. Lane13- is negative blank control.
Dendrogram of random amplified polymorphic DNA (RAPD) profile based on UPGMA (Unweighted pair group mathematical average clustering algorithm).
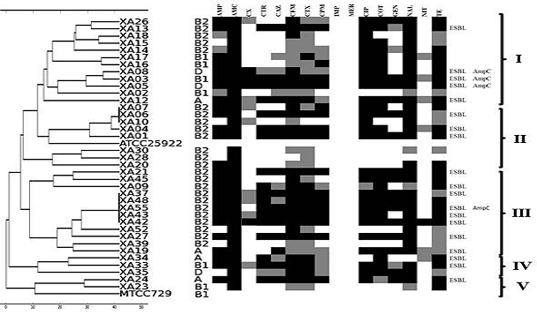
RAPD analysis revealed the genetic correlation among the isolates. The isolates were divided into 5 different clusters. Cluster I, IV and V were mixed clusters, which included the phylogenetically distinct isolates. The isolates from a single phylogenetic group were closely related with some exceptions. Cluster II and III were full of B2 isolates with an exception of XA19, group A isolate in cluster III. The antibiotic resistant profile of the isolate correlated with the RAPD profile. The multiple drug resistant and ESBL producing isolates were distributed widely in all the clusters.
Discussion
Worldwide E. coli is the more common bacteria present in UTIs including India [7,20,21]. The current concern of UTIs is the prevalence of multi drug resistance and also a major problem in the treatment of UTIs [21,22]. Generally the bacterium E. coli may be attaching itself to the cell lining of the urinary tract and produces a protect film that might be the cause of resistant to medication. In this study most of the E. coli (28 strains) were positive for ESBL out of which seven isolates also produced AmpC β-lactamases. The incidence of UTI was higher among female (59%) and their infections tend to recurrent compared to male patients (41%).
In the present study UTI is quite common among female (26%) belonging to the age group 11-30 years and also elder male (19%) of above 51 years. Earlier reports from India and other countries represented the same [22,23] also supported the results as E. coli infection was higher among female than male. However earlier reports indicated that UTIs were more common among female belonging to the age group of 21-30 years [24,25]. Battacharyya et al., reported that the mean age of female having UTIs was 31.1 years [26]. Kumar et al., reported that UTIs were more common among male (33.3%) belonging to age group 51-75 than that of female (18.9%), whereas among children (below 10 years) UTIs were found only among female [25]. Our results also correlated with this with higher level of infection among elderly male. Segar et al., reported that among female, UTIs were more common among age group 11-40 years, whereas among elderly male with above 51 years were more susceptible to UTIs [27]. Dash et al., reported that UTIs were more common among young female (18-27 years) and elderly male (≥68 years). Urinary incontinency and poor hygienic practice in elder people were related to cause of UTI [28].
Resistance to extended spectrum cephalosporins due to the production of ESBL and/or AmpC β-lactamase was being reported recent years in India [29,30]. Both ESBL and plasmid mediated AmpC β-lactamases are associated with multidrug resistance. Phenotypic detection method for ESBL is based on the resistance to cephalosporins, whereas for AmpC is based on the resistance to cephamycins [31]. In this study seven isolates showed resistance to both cephalosporins and cephamycins. Phenotypic detection method indicated the presence of both ESBL and AmpC β-lactamases in all the isolates. Higher amount of resistance (84-93%) to nalidixic acid was reported already from several areas globally [25,32,33]. Resistance to amoxicillin/clavulanic acid was increasing recent years. Mittal et al., reported 95%, Datta et al., reported 88.57% resistance to amoxicillin/clavulanic acid, where as 100% resistance was found among our isolates [24,34]. It is not a new thing that the evolution of resistance among antibiotics. However the periodic observation of antibiotic profile and its resistance will help us to treat UTI infection.
The presence of intestinal or commensal group A strains in the UTIs indicates the same, that the faecal contamination may be the source of infection. Among male the age group acquired higher infection. Sabate et al., reported that the commensal intestinal flora (group A and B1) of the patients with UTI were more virulent than the same phylogenetic groups isolates from the normal individuals [35]. The higher prevalence (33%) of group A isolates in our study indicates the essence of attention needed on the commensal flora. The selective pressure due to antibiotic usage in their ecological niche results the spread of ESBL isolates in extraintestinal environments [36]. It was also found that the prevalence of highly virulent phylogenetic group B2 (41%) among the ESBL isolates in Tamil Nadu. The previous findings also suggested that the highly pathogenic nature of the group B2 might be one of the reasons to cause UTI.
So the intestinal tract acts as the reservoir for the uropathogens of group A and B2. All the isolates in this study were susceptible to carbapenems. However, the geographical variation and infrequent usage of carbapenems may be the cause of infrequent isolation of NDM strains. This also correlated to the findings of Hussain et al., [5].
Worldwide spreading of E. coli belonging to different clonal groups was one of the major problems in the epidemiology of UTIs. The global spread of CTX-M15 producing multidrug resistant ST131 strains among the urinary tract infections in community setting has been reported. In India also the wide spread CTX-M15 producing E. coli strains has been reported since 2006 and it was the only dominant enzyme distributed among various strains [37]. In early 2011, E. coli ST131 was reported and since then several studies have been carried out [38]. However, the antibiotic resistance pattern of community acquired UTI Escherichia coli from local area had not been analysed with a set of antibiogram profiling. In our present study, we have analysed and found that 18 out of 58 strains (31.03%) as ST131, out of which 10 strains were ESBL (17.24%).
Limitation
The outcome results of this study had still some limitations. We have considered urinary tract infection of E. coli and not other microbes with irrespective of age and sex. The sample size may be increased in future considering inpatients in hospital to get more useful information.
Conclusion
From this study, we noted phenotypically most of the strains were positive for ESBL and showed high sensitivity for Nitrofurantoin and cefoxitin. Further studies related to MLST and whole genome sequencing of selected isolates from different MLST clades will be useful to understand the genetic diversity of UTI from India.