With the increase in biofilm-related infection, researchers have tried to examine new ways to prevent and remove microbial biofilms from an infected site to control the surface attached bacteria. Among the various strategies have been proposed to control biofilm, one of them being using “nonantibiotic drugs” for their antimicrobial or antibiofilm activities [8]. One of the such examples is coating various medical devices with EDTA aimed at inhibiting initial cell adhesions [9].
Compounds like sodium ethylenediaminetetraacetate (EDTA) posses some antimicrobial activity, the potency of which can be increased by combining other molecules [10] or antimicrobials along with EDTA [11,12]. It should be mentioned here that cations are required to stabilize the negatively charged polysaccharides which is required to hold the biofilm together EDTA forms strong complexes with these cations thereby chelating them and destabilizing the formed biofilm [13]. In the present study, we have tried to find out the inhibitory effect of EDTA against MRSA strains with a capacity to form the biofilm.
Materials and Methods
The clinical isolates of MRSA and its microbiological testing: After getting ethical clearance from the institutional Ethical Committee of Medical College and Hospital, Kolkata quasi Experimental (nonrandomized, pre-post-intervention) study was performed on the 25 strong biofilm forming clinical strains isolated in the Microbiology Department, Medical College, Kolkata from Jan 2013 to Dec 2013. Firstly, Staphylococcus aureus strains were isolated from the different samples (pus, sputum, urine, blood, fluid, and other miscellaneous samples like central venous catheter tip, endotracheal tube, orthopedic implants, etc) of the patients received by the Microbiology Department for routine examination. The samples were initially streaked in Blood agar plates followed by 24 hours incubation at 37oC. Staphylococcus aureus was confirmed by Gram staining, slide coagulase test, tube coagulase test and anaerobic mannitol fermentation by using standard methods [14]. All the confirmed Staphylococcus aureus strains were further screened for methicillin resistance with the help of cefoxitin discs (30 μg/disc) (Hi-Media, India) by using Kirby-Bauer disc diffusion method. The isolates were considered resistant if the zone of inhibition was 21mm or less (10). Followed by other antibiotics like Clindamycin (2μg) Erythromycin (15 μg), Vancomycin (30 μg), Amikacin (30μg), Gentamycin (10μg), Cefuroxime (μg), Levofloxacin(5 μg), Ciprofloxacin (5 μg), Linezolid (30 μg), Doxycycline (30 μg), Amoxycillin (10μg), Amoxyclav (30μg)on Mueller-Hinton agar. These antibiotic discs were obtained from commercial source. Results were interpreted according to CLSI guidelines (2014) [15]. Throughout the study S. aureus ATCC 25923 was used as a control.
Bacterial DNA isolation: Genomic DNA was isolated with phenol and chloroform method and was precipitated with ethanol by standard methods [16]. The only addition was lysostaphin (40 mg per ml) and incubation for 30 min at 37°C prior to addition of RNase. The DNA was recovered by centrifugation, vacuum dried and finally dissolved in 200μl Tris- EDTA buffer (10mM Tris, 1mM EDTA (pH 8.0), and concentration was determined spectrophotometrically at 260nm. An amount of 10ng template DNA was used in this study.
PCR primer design and amplification: By using specific accession numbers nucleotide sequences, each of the desired genes were obtained from GeneBank. The identities of S. aureus isolates were confirmed by species-specific gene amplification (16S rRNA) and the presence of mecA gene, responsible for methicillin resistance was carried out by employing the sets of gene specific primers in PCR assay. For the PCR amplification, the reaction mixture composition was similar for both the genes and oligonucleotide primers used in the study. DNA amplification was performed in 25μl of reaction mixture that contained 2.5 μl l0x buffer, 2.5 μl of 200mM dNTP mix (I.D.T), 2 μl of 25 mM of MgCl2, 1.5μl each of a pair of primers forward and reverse (10 pmol/μl, I.D.T), 10 ng of staphylococcal template DNA and 1 U of Ampli-TechgoldTaq DNA polymerase. PCR amplification was carried out in a DNA thermal cycler (Applied Biosystem 2720) under reaction conditions as described. Denaturation, annealing and extension temperatures were kept at 94°C, 64 °C (for 16S r RNA), 54 °C (for mecA) and 72oC respectively for each gene amplification [17,18].
The amplification product was resolved by electrophoresis on 1.5 % agarose gel at 70V (constant voltage) and visualized with ultraviolet light after staining with ethidium bromide. Product size was determined by using the 500bp DNA molecular weight ladder (NEB).
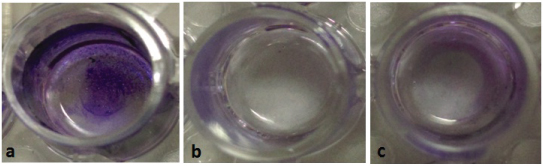
Biofilm formation and its treatment: For detection of biofilm we used quantitative microtitre plate method [19]. Isolated colonies of MRSA strains were inoculated in 5ml of trypticase soy broth (TSB) with 1% glucose and then it was incubated at 37°C for 24 h. The cultures were then diluted with fresh medium at ratio of 1:100. Each well of sterile 96 well-flat polystyrene tissue culture treated plates (Tarson, India) was inoculated with 200 μL of the diluted broth cultures of different MRSA strains isolated from various samples. S. aureus ATCC 25923 was used as positive biofilm control. Negative control wells were inoculated with sterile broth. The plates were incubated at 37°C for 24 h. After incubation, the wells were washed with 0.2 ml of phosphate buffer saline (PBS) (pH 7.2) four times. This removed free floating bacteria. Biofilm formed by bacteria adherent to the wells were fixed by keeping in hot air oven at 120°C for 60 minutes and were then stained by crystal violet (CV) (0.1%). Excess stain was removed by using deionized water by rinsing four times with sterile distilled water and subsequent decolorizing with 30% acetic acid. Optical density (OD) of stained adherent biofilm was obtained by using micro ELISA autoreader (model 680, Biorad, UK) at wavelength 570 nm. Uninoculated wells containing TSB were used as blanks. Blank-corrected absorbance values were used for reporting biofilm production. The test was performed in triplicate and repeated thrice [Table/Fig-1a]. The biofilm formation was interpreted according to the criteria laid down by Stepanovic et al., [20].
< 0.120 Non Biofilm producer.
0.120 – 0.240 Moderate Biofilm producer.
> 0.240 Strong Biofilm producer.
Inhibition by EDTA treatment: The method of Amalaradjou et al., was used to assess the ability of the EDTA to inhibit MRSA biofilm production [21]. MRSA strains were separately grown overnight in TSB at 37°C.
Two hundred microliters of this culture were used as the inoculum (~6.0 log CFU) and transferred into sterile 96-well polystyrene tissue culture plates, followed by the addition of 0 (negative control), 0.5 (0.2 μl), 1 (0.4 μl), 2 (0.8 μl)and 4(1.6 μl) mM of EDTA. The plates were incubated at 37°C for 24 hrs and washed four times with 200 μL of sterile PBS, dried and stained with 1% crystal violet for ELISA reading. After rinsing, optical density (OD) of stained adherent biofilm was obtained by using micro ELISA auto reader at wavelength 570 nm. Wells containing TSB without EDTA were used as blank. Triplicate samples were included for each treatment, and the experiment was replicated three times.
Reduction of biofilm by EDTA treatment: To evaluate the ability of the EDTA to dissociate MRSA biofilm 200μL of the each bacterial cell suspension (~6.0 logCFU) were inoculated in sterile 96-well polystyrene tissue culture plates and incubated at 37°C for 24 h without agitation for biofilm production [20]. The overnight aggregated culture (formed biofilm) was then exposed for 24 hours with different concentrations 0 (negative control), 2.5 (1 μl), 5 (2μl),10 (4 μl) and 20 (8 μl) mM of EDTA by adding it to the microtitre plate. Thereafter, the wells were washed four times with 200 μL of sterile PBS, dried and stained with 1% crystal violet for ELISA reading. After rinsing, the absorbance of the adherent biofilm was measured at 570 nm in a microplate reader. Wells containing TSB without EDTA were used as control. Triplicate wells were included for each treatment.
Determination of Bacterial Counts and Sensitivity pattern in Biofilm: The antibiofilm effect of EDTA was studied by counting surviving bacterial populations in the biofilm by viable plate count method [21]. Following treatment with EDTA, the wells were washed four times with PBS, and the adherent biofilm was scraped and plated on Nutrient Agar. The plates were incubated at 37°C for 24 h before enumerating the bacterial colonies. The isolated bacterial colonies were then subsequently tested for methicillin and other antibiotic resistance by Kirby-Bauer disc diffusion method discs on Mueller-Hinton agar. The plates were incubated at 370C for 24 hours and zone size was recorded. Minimum Inhibitory Concentration (MIC) for methicillin was also determined by using Ezy MIC strip according to the instructions given by the manufacturer (Hi Media Laboratories, India).
Statistical Analysis
The t-test was utilized for calculating the difference of mean between OD value for control (0mM EDTA) and different concentration of EDTA used for both reduction and inhibition. The results of biofilm inhibition and reduction were significantly lower (p < 0.00) for all the different concentrations of EDTA.
Results
Samples were collected in the Microbiology Department of Medical College from patients coming to the hospital during the year 2013 in West Bengal, India. Gram positive bacterial pathogen was identified in 2987 samples of which 420 (14.06%) strains were that of Staphylococcus aureus. Out of which 96 (22.8 %) were MRSA isolates. From this 25 (26.04%) were strong biofilm formers.
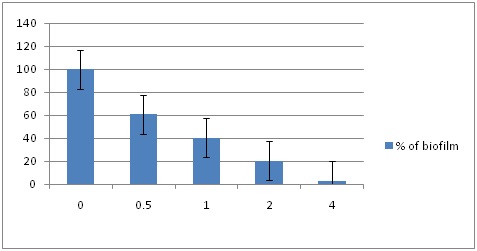
Biofilm Inhibition: To assess the ability of EDTA to inhibit biofilm, 25 MRSA samples were loaded in triplicate in 96-well plates in presence of different concentrations of EDTA i.e. 0, 0.5, 1, 2 and 4mM. As seen by crystal violet biofilm quantification at 570nm, significant inhibition (96.96%) was observed in the wells with 4Mm concentration for each of 25 MRSA samples [Table/Fig-1b,2].
Biofilm reduction and inhibition assay. (a. pre-formed biofilm) MRSA biofilms were developed on microtitre plates for 24 hrs, (b. biofilm inhibition) after addition of different concentrations of EDTA and (c. biofilm reduction) after treatment of biofilm with different concentrations of EDTA. Biofilms were rinsed and stained with CV.
Quantification of biofilm inhibition formed on microtitre plate after treating with different concentrations (mM) of EDTA in each of 25 MRSA samples as shown in Fig. Each experiment was carried out three times, yielding almost similar results. Data are represented as the mean ± SEM.
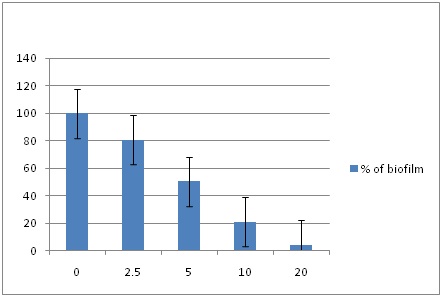
Biofilm Reduction: EDTA was also effective at removing fully formed biofilms of MRSA on polystyrene plates. The 96-well microtitre plates with fully formed biofilm in 25 MRSA samples was treated with different concentrations i.e. 0 (Control), 2.5, 5, 10, 20 mM of EDTA. As seen by crystal violet biofilm quantification at 570nm, significant reduction of biofilm was observed in the wells for each of 25 MRSA samples. EDTA at 20 mM concentration for 24 hrs of exposure, showed almost complete reductionof biofilm (95.5%) of the MRSA strains [Table/Fig-1c,3].
Reduction of biofilm on microtitre plate after treating with different concentrations (mM) of EDTA in each of 25 MRSA samples. Each experiment was carried out three times, yielding almost similar results. Data are represented as the mean ± SEM.
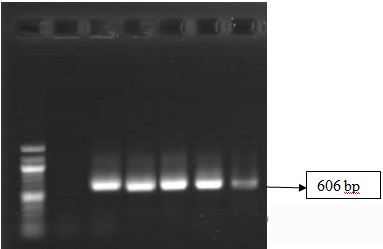
Conversion of MRSA into MSSA: The simple viable plate count method helped to demonstrate the surviving bacterial populations in the biofilm. From these bacterial populations, an inoculum was prepared for antimicrobial susceptibility test by Kirby Bauer Disc Diffusion Method. After treatment with EDTA, the MRSA strains which were resistant to all the drugs as mentioned above became sensitive to those drugs including cefoxitin. EDTA, thus converted MRSA strains to MSSA strains. MIC for cefoxitin was again determined by using Ezy MIC strip according to the instructions given by the manufacturer. MIC was found to be ≤22μg/mL confirming the conversion of MRSA isolates to MSSA, after treatment with 10mM concentration of EDTA. Further to test whether any genetic change was introduced in EDTA treated MRSA strains, the presence of mecA gene was again analyzed by PCR. But the PCR amplification revealed the presence of mecA genes in genomic isolates, confirming only the phenotypic changes [Table/Fig-4].
Agarose gel electrophoresis of PCR-amplified mecA genes at 606 bp of EDTA treated MRSA isolates (i) Lane 1: 1000 bp ladder; Lane 2: Negative Control; Lane 6: Positive Control S. aureus ATCC 33591 (ii) Lane 3-5: MRSA isolate after EDTA treatment at 2.5, 5, 10 and 20 mM conc
Discussion
Sometimes, the various biomedical devices used for different clinical procedures may be associated with infections due to microbial colonization and multiplication [22]. Among them, Staphylococci mainly causes multiple infections on foreign devices like orthopedic and surgical implants, stents, intravenous catheters, infusion pumps, mechanical heart valves, pacemakers, surgical implants etc where biofilms can results in serious complications leading to high morbidity, mortality, and medical costs [23]. For eradicating the biofilm from the infected site it usually requires surgical intervention with antimicrobial therapy [24]. The bacteria enclosed in a biofilm layer are usually multi-drug resistant compare to free floating form. Therefore, a possible therapeutic approach to biofilm associated medical device infections involves the use of EPS-degrading agents that might reduce or remove cell-to-cell and cell-to-surface associations within the biofilm, thus releasing the planktonic cells into the environment and allowing the host immune response and antimicrobial agents to act easily upon them [25,26]. In this study, we examined the potential use of EDTA as a biofilm control agent against MRSA. It has already been reported in some of the literature that EDTA, a chelator of calcium and magnesium, and an anticoagulant, possesses potential activity against planktonic microbial cells and inhibits their biofilms [27,28]. Root et al., also demonstrated the effect of EDTA on biofilm formed by S. epidermidis (invitro) on a Hickman catheter made of silicone material. They concluded that a high concentration of EDTA i.e. 108 mmol/l could eradicate the staphylococcal biofilm [28]. In one of the studies, EDTA treatment with 40 mg/ ml for 24 hrs significantly reduced the viability of biofilms of S.epidermidis, MRSA, P. aeruginosa, K. pneumoniae or C.albicans [29]. On the other hand, according to some authors higher than 2 mmol/l concentrations of EDTA, was in vitro toxic for viability of microbial cell cultures [30]. In another study conducted by Marek et al., EDTA inhibited formation of biofilm in S. epidermidis on medical devices at 1.0–2.0 mmol/l concentration and reduced the mature biofilm in some strains at 2.0–4.0 mmol/l EDTA whereas for the other isolates, EDTA (>32 mmol/l) was required at higher concentrations [31]. But one of the studies showed no significant decrease in Staphylococcus biofilm, even after an exposure to the EDTA for 60 min [32]. EDTA may also be combined with other molecules like minocycline, ovotransferin, protamine sulfate or antibiotics for a synergistic antibiofilm effect against Staphylococcus spp [33]. Another recent study indicated that more than 1.5 mg/ml of EDTA inhibited the growth of L. monocytogenes. However, when EDTA was combined with ovotransferrin, the strength of its action decreased [34]. Our results showed that EDTA at 4mM and 20 mM concentration for 24 hours, demonstrates a significant inhibition and reduction of biofilm of the MRSA strains respectively. EDTA has been shown to lead to phenotypic mutation by converting MRSA into MSSA. This could be probably due to the fact that EDTA selectively solubilizes EPS membrane and increases membrane permeability of MRSA, allowing the antibiotics to penetrate inside thereby reducing methicillin resistance. Supporting this there are some reports stating that the inhibitory effect of EDTA on staphylococcal biofilm formation is due to the fact that EDTA causes the chelation of metal ions [33]. The appropriate biofilm surface properties are determined by the presence of PIA (Polysaccharide Intracellular Antigen) which in turn depends on the Mg2+ ions level [35]. Therefore, the decreased level of Mg2+ ions (which is mainly due to chelation by EDTA) inhibits the adhesion process, thereby inhibiting the biofilm formation [33].
Since MRSA have the ability to persist in the hospital environment and form biofilms on a wide variety of biotic and abiotic surfaces, EDTA can be used as potential therapeutic and prophylactic antimicrobial agent. With the help of aseptic techniques and proper utilization of EDTA solutions, foreign body associated infections can be reduced, thus decreasing the morbidity and mortality rate.
Conclusion
In conclusion, our study demonstrates that EDTA is effective in preventing biofilm formation by MRSA and inactivating pre-formed biofilms on polystyrene plates. These results suggest that EDTA can be potentially used as a sanitizer for hospital surfaces. However, further experiments are needed to evaluate the efficacy of EDTA in comparison with other anti-MRSA therapies both invitro and in vivo. In future, these findings would be beneficial in our battle against superbugs.
Conflict of Interests: The authors declare that there is no conflict of interests regarding the publication of this paper.