Prevalence of MRSA Nasal Carriage in Patients Admitted to a Tertiary Care Hospital in Southern India
Kalpana George1, Jasmine Kulapurathu Abdulkader2, Madhan Sugumar3, Girija Kalarikkal Rajagopal4
1 Assistant Professor, Department of Microbiology, Government Medical College, Kozhikode, Kerala, India.
2 Assistant Professor, Department of Microbiology, Government Medical College, Thrissur, Kerala, India.
3 Research Fellow, Department of Microbiology, JIPMER, Puducherry, India.
4 Professor, Department of Microbiology, Amrita Institute of Medical Sciences, Cochin, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Kalpana George, Assistant Professor, Departmen of Microbiology, Government Medical College, Kozhikode, Kerala- 673008, India. E-mail : Kalpana.george@gmail.com
Introduction
Infections with MRSA, both community and hospital acquired, are well established and the source of infection is often a carrier. There are very few studies showing the magnitude of MRSA nasal colonization among healthy persons from the community. This study was conducted to detect the prevalence of MRSA nasal carriage in patients who did not have any known risk factors associated with HA- MRSA colonization, admitted to a tertiary care centre in Kerala.
Materials and Methods
Nasal swabs were collected from patients within 24 hours of admission. Specimen were inoculated on chromogenic agar (HiCrome MeReSa agar-HiMedia) for MRSA screening. Isolates were then subjected to antibiotic sensitivity tests, SCCmec typing and PVL gene detection.
Results
Out of 683 patients, 16 carried MRSA in their nares (2.3%). Of the 16 strains 13 (81.25 %) strain were SCCmec type III and one belonged to SCCmec type IV (6.25 %). Two strains failed to amplify SCCmec genes. Three strains carried genes for PVL toxin (18.75%).
Conclusion
With a better understanding of the complex epidemiology of MRSA it is increasingly apparent that demarcations between the HA and CA phenotypes are not as clear cut as previously thought. In this study of nasal carriage of MRSA in the community we have demonstrated prevalence consistent with published data. Most isolates however were shown to belong to the type conventionally assigned to HA-MRSA.
Nasal colonization, Nasal swabs, Phenotypes
Introduction
Methicillin resistant Staphylococcus aureus (MRSA) has gained much interest since it is first reported in 1961 and it has become epidemic in many hospitals across the globe. In India the rate of nosocomial infections with MRSA is around 40% [1,2].
MRSA has ceased to be a hospital only pathogen. Over the last two decades they have gained importance as community acquired pathogens. Healthcare (HA) associated and community (CA) associated MRSA strains can be distinguished genetically. Hospital acquired MRSA (HA-MRSA) carries staphylococcal cassette chromosome mec (SCCmec) type I, II and III and community acquired S.aureus (CA-MRSA) carries SCCmec types IV, V or VI and genes encoding for Panton-Valentine leukocidin (PVL) [3].
HA-MRSA infections are associated with risk factors like recent hospital stay, living in a long term care facility or carrying an indwelling device or catheter. However, individuals may be colonized with HA–MRSA without these risk factors. CA-MRSA infection and colonization are not associated with the risk factors mentioned above. It is now recognised that CA-MRSA is as important as HA-MRSA in causing hospital outbreaks [4].
The magnitude of nasal colonization of MRSA in personnel who come into close contact with health care facilities is well established. There is however, a paucity of data from healthy individuals in the community. Published data from various centres in India project a prevalence of 1- 5% [5–9].
We present here a study conducted to document the prevalence of MRSA nasal carriage in patients who did not have any known risk factors associated with HA-MRSA colonization, admitted to a tertiary care centre in Kerala. We hypothesised that if at all these patients carry MRSA in their nares, it would be CA-MRSA. We also aimed at finding out the SCCmec type and presence of PVL gene among the MRSA isolates.
Materials and Methods
This was a cross-sectional study conducted in the Department of Microbiology, Government Medical College, Thrissur, Kerala between April 2012 and May 2013. Approval from Institutional Ethics Committee was obtained. Informed consent was taken from patients before swabbing.
The minimum sample size was calculated as 475, anticipating a prevalence rate of 5% based on previous studies and to detect the prevalence with 2% precision. The study subjects were adult patients admitted to surgical, orthopaedics and subspeciality wards within 24 hours of admission.
The following categories of patients were excluded from the study.
1. Patients admitted in Intensive Care Units and pay wards.
2. Patients with any of the following history were excluded from the study:
- Hospitalisation within past 3 months.
- History of antibiotic used within past 3 months.
- Being in close proximity to other ill patients in hospital or household.
Colonization of MRSA is defined as isolation of MRSA in the nasal swabs in the absence of systemic signs of sepsis or overt infection.
Specimens were collected from the anterior nares using cotton swabs moistened with glucose broth. The walls of the vestibules of anterior nares were thoroughly swabbed taking sufficient time. The swabs were inoculated into brain heart infusion broth, incubated for 6-8 hours and subcultured onto chromogenic agar (HiCrome MeReSa agar-HiMedia) and incubated for 48 hours with reading at the end of 48 hours. Suspected colonies from chromogenic agar were confirmed as MRSA by grams stain, growth on mannitol salt agar, catalase and coagulase tests as per standard guidelines. Commercially available discs were used for antibiotic sensitivity testing by Kirby Bauer method on Muller Hinton agar according to performance standards of Clinical and Laboratory Standards Institute (CLSI) [10]. The antibiotics tested were Penicillin (10 U), Erythromycin (15μg), Gentamycin (10μg), Cephalexin (30μg), Cefoxitin (30μg) and Cotrimoxazole (1.25-23.75μg). Zone sizes were interpreted according to CLSI guidelines.
The strains were further subjected to PCR at Christian Medical College, Vellore to
- Confirm the presence of mec A gene
- Type the SCCmec
- Detect the presence of PVL gene
PCR for the detection of SCCmec and PVL gene was done according to procedure guidelines by Oliveira DC et al., and Lina G et al., respectively [11,12].
Results
A total of 683 patients fulfilled the criteria to be included in the study. Out of 683 patients, 16 carried MRSA in their nares (2.3%).
They were resistant to all the antibiotics tested viz Penicillin (10 units), Erythromycin (15μg), Gentamycin (10μg), Cephalexin (30 μg), Cefoxitin (30 μg) and Cotrimoxazole (1.25-23.75 μg).
[Table/Fig-1,2] show PCR control runs and interpretations for SCCmec and PVL genes respectively.
Control run and interpretation of SCCmec gene typing.
| S.No | Strain ID | Interpretation |
|---|
| 1 | M.H20 (Negative control) | Satisfactory |
| 2 | Ladder 100bp | Satisfactory |
| 3 | Control strain 1 | Type-III A |
| 4 | Control strain 2 | Type-III |
| 5 | Control strain 3 | Type-III |
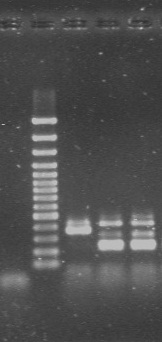 |
Control run and interpretation of PVL gene PCR.
| S.No | Strain ID | Interpretation |
|---|
| 1 | 9D13 | Negative (old primer) |
| 2 | GRE (Control) | Negative (old primer) |
| 3 | 1D30 | Negative (old primer) |
| 4 | Ladder | 100bp |
| 5 | M.H20 (Negative control) | Negative |
| 6 | Test strain (1D30) | Negative |
| 7 | GRE | PVL Positive 433bp |
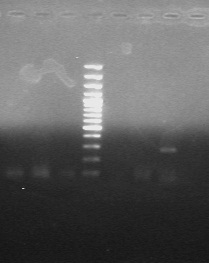 |
Of the 16 isolates 13 (81.25 %) strains were SCCmec type III and one belonged to SCCmec type IV (6.25%). Two strains failed to amplify the SCCmec genes. Three strains carried genes for PVL toxin (18.75%) [Table/Fig-3].
Results of SCCmec typing and PVL gene PCR.
| Test strain | SCC mec gene | PVL gene | Test Strain | SCCmec gene | PVL gene |
|---|
| Test strain 1 | Type-III | Negative | Test strain 9 | Type-III A | Negative |
| Test strain 2 | Type-III | Negative | Test strain 10 | Type-III B | Negative |
| Test strain 3 | Type-III | Negative | Test strain 11 | Type –III B | Positive(433bp) (433bp) |
| Test strain 4 | Type-III | Negative | Test strain 12 | Type –III | Positive(433bp) |
| Test strain 5 | Type-III | Negative | Test strain 13 | Negative | Negative |
| Test strain 6 | Type-III | Negative | Test strain 14 | Type –IV | Negative |
| Test strain 7 | Type -III | Negative | Test strain 15 | Negative | Negative |
| Test strain 8 | Type –III | Negative | Test strain 16 | Type –III | Positive(433bp) ((((433bp) |
Discussion
We studied the rates of MRSA colonization of anterior nares in adults within 24 hours of admission to a tertiary health care hospital in Southern India.
We chose to swab the anterior nares as they are the natural niches of S.aureus and are the most consistent areas from which the organism has been isolated [13,14]. Rectum, perineum, axilla and device insertion sites are the other favoured sites for colonization. However these sites were excluded anticipating that fewer patients would likely consent for participation.
Isolation rates of S.aureus and MRSA from our routine clinical specimens in our laboratory has been approximately 8% and 4% respectively. In the present study, prevalence of MRSA colonization was found to be 2.3%. These patients did not have history of recent hospital admission or other risk factors associated with MRSA colonization. The nasal swabs were collected within 24 hours of admission to exclude colonization of nares resulting from hospitalization.
The prevalence of MRSA colonization is comparable to other published data from India. In a study conducted by Pathak et al., among healthy preschool children in Ujjain showed prevalence of MRSA nasal colonization as 1.02% [5]. Nasal carriage rates of MRSA were found to be 1.83% and 3.89% from similar studies conducted at Kashmir and Chandigarh respectively [6,7]. Colonization rates of MRSA outside hospital environments have been documented to be low across geographic areas [15–18].
All 16 isolates in our study were found to be resistant to all antibiotics tested, a finding distinct from other published Indian studies [6,7]. The potential of these colonizers to act as sources for hospital infection outbreaks with multidrug resistant strains is a cause for concern.
We hypothesized that most of the isolates would belong to SCC mec type IV, V or VI. Interestingly in our study the majority (13/16) of isolates were found to carry SCCmec type III and only three had PVL gene. In a study conducted in Taiwan on 6057 children between 2 -5 years in a community setting, it was found that 18% of MRSA carriers had SCCmec type II and III in their genomes and PVL gene was absent [19]. In a population based study conducted in Brazil to detect nasal colonization of MRSA, it was found that all MRSA isolates belonged to SCCmec type IV and none of them had PVL gene [20]. It has been recognised that presence of PVL alone does not predict the community origin of MRSA colonization [21]. However, the large population based NHANES study showed a specific association of SCCmec type IV with PVL gene. 50.7% of MRSA were SCCmec type IV and 49.3% SCCmec type II in this study [22]. In India, SCCmec type III is the most prevalent strain in hospital setting and there is insufficient data regarding SCCmec types prevalent in the community [23].
The distinction between CA-MRSA and HA-MRSA isolates has been found to have blurred recently. Increasing cross transmission of HA and CA-MRSA between hospitals and communities has rendered the epidemiology of MRSA complex. Centres for Disease Control and Prevention investigators have proposed a category of MRSA infections, “health care-associated, community-onset” MRSA (HACO-MRSA) infection which includes cases that would be HA-MRSA infections by history of health care exposure but have onset in the community [24]. The colonizing strains that we isolated were likely to be acquired from the community, but showed a pattern suggestive of origin in the hospital.
Limitation
We have identified some limitations in our study. Sites other than anterior nares were not swabbed and this may have resulted in underestimation of MRSA colonization. Nasal colonization of S.aureus and MRSA is dynamic in nature and hence it is possible to have ‘intermittent carriage states’. Cross sectional design of this study makes predictions of changes in colonization with time impossible. Epidemiological typing was not done; hence clonal origin of the organism could not be established.
Conclusion
With a better understanding of the complex epidemiology of MRSA it is increasingly apparent that demarcations between the HA and CA phenotypes are not as clear cut as previously thought. In this study of nasal carriage of MRSA in the community we have demonstrated prevalence consistent with published data. Most isolates however were shown to belong to the type conventionally assigned to HA-MRSA. Further studies are required to characterise MRSA types in the community and more importantly, to identify whether a true redistribution of the HA-MRSA strains is occurring.
[1]. Rallapalli S, Verghese S RV, Validation of multiplex PCR for simultaneous detection, identification of methicillin resistant Staphylococcus aureusIndian J Med Microbiol 2008 28:82-83. [Google Scholar]
[2]. Joshi S, Ray P, Manchanda V, Bajaj J, Chitnis DS, Gautam V, Methicillin resistant Staphylococcus aureus (MRSA) in India: prevalence & susceptibility patternIndian J Med Res 2013 137(February):363-69. [Google Scholar]
[3]. David MZ, Daum RS, Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemicClin Microbiol Rev 2010 23(3):616-87. [Google Scholar]
[4]. Schinasi L, Wing S, MacDonald PDM, Richardson DB, Stewart JR, Augustino KL, Medical and Household Characteristics Associated with Methicillin Resistant Staphylococcus aureus Nasal Carriage among Patients Admitted to a Rural Tertiary Care HospitalPLoS ONE 2013 8(8) [Google Scholar]
[5]. Pathak A, Marothi Y, Iyer RV, Singh B, Sharma M, Eriksson B, Nasal carriage and antimicrobial susceptibility of Staphylococcus aureus in healthy preschool children in Ujjain, IndiaBMC Pediatr 2010 10(1):100 [Google Scholar]
[6]. Fomda BA, Thokar MA, Khan A, Bhat JA, Zahoor D, Bashir G, Nasal carriage of Methicillin-resistant Staphylococcus aureus among healthy population of Kashmir, IndiaIndian J Med Microbiol 2014 32(1):39-43. [Google Scholar]
[7]. Chatterjee SS, Ray P, Aggarwal A, Das A, Sharma M, A community-based study on nasal carriage of Staphylococcus aureusIndian J Med Res 2009 130(6):742-48. [Google Scholar]
[8]. Chande CA, Shrikhande SN, Jain DL, Kapale S, Chaudhary H PR, Prevalence of methicillin-resistant Staphylococcus aureus nasopharyngeal carriage in children from urban community at NagpurIndian J Public Health 2009 53(3):196-98. [Google Scholar]
[9]. Govindan S, Maroli AS, Ciraj AM, Bairy I, Molecular epidemiology of methicillin resistant staphylococcus aureus colonizing the anterior Nares of school children of Udupi TalukIndian J Med Microbiol 2015 33(Suppl):129-33. [Google Scholar]
[10]. CLSI-Performance standards for antimicrobial susceptibility testing: twenty-second informational supplement. Wayne, PA: Clinical and Laboratory Standards Institute; 2012 [Google Scholar]
[11]. Oliveira DC, De Lencastre H, Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureusAntimicrob Agents Chemother 2002 46(7):2155-61. [Google Scholar]
[12]. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumoniaClin Infect Dis Off Publ Infect Dis Soc Am 1999 29(5):1128-32. [Google Scholar]
[13]. Garza D, Sungar G, Johnston T, Rolston B, Ferguson JD, Matheson GO, Ineffectiveness of surveillance to control community-acquired methicillin-resistant Staphylococcus aureus in a professional football teamClin J Sport Med Off J Can Acad Sport Med 2009 19:498-501. [Google Scholar]
[14]. Kluytmans J, Belkum AV, Verbrugh H, Nasal carriage of Staphylococcus aureus : Epidemiology , Underlying Mechanisms , and Associated RisksClin Microbiol Rev 1997 10(3):505-20. [Google Scholar]
[15]. Sharma Y, Jain S, Singh H, Govil V, Staphylococcus aureus: Screening for Nasal Carriers in a Community Setting with Special Reference to MRSAScientifica 2014 2014:479048 [Google Scholar]
[16]. Olsen K, Sangvik M, Simonsen GS, Sollid JUE, Sundsfjord A, Thune I, Prevalence and population structure of Staphylococcus aureus nasal carriage in healthcare workers in a general population. The Tromsø Staph and Skin StudyEpidemiol Infect 2013 141(1):143-52. [Google Scholar]
[17]. Melles DC, Tenover FC, Kuehnert MJ, Witsenboer H, Peeters JK, Verbrugh HA, Overlapping population structures of nasal isolates of Staphylococcus aureus from healthy Dutch and American individualsJ Clin Microbiol 2008 46(1):235-41. [Google Scholar]
[18]. Mehraj J, Akmatov MK, Strömpl J, Anja Gatzemeier FL, Guido Werner, Dietmar H, Pieper, Eva Medina, Wolfgang Witte, Frank Pessler G rard K. Methicillin-Sensitive and Methicillin-Resistant Staphylococcus aureus Nasal Carriage in a Random Sample of Non-Hospitalized Adult Population in Northern GermanyPLoS One 2014 24(9)(9):e107937 [Google Scholar]
[19]. Chen CJ, Hsu KH, Lin TY, Hwang KP, Chen PY, Huang YC, Factors associated with nasal colonization of methicillin-resistant Staphylococcus aureus among healthy children in TaiwanJ Clin Microbiol 2011 49(1):131-37. [Google Scholar]
[20]. Pires FV, da Cunha M, de LR, de S, Abraão LM, Martins PYF, Camargo CH, Fortaleza CMCB, Nasal carriage of Staphylococcus aureus in Botucatu, Brazil: a population-based surveyPloS One 2014 9(3):e92537 [Google Scholar]
[21]. Rossney AS, Shore AC, Morgan PM, Fitzgibbon MM, O’Connell B, Coleman DC, The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in IrelandJ Clin Microbiol 2007 45(8):2554-63. [Google Scholar]
[22]. Kuehnert MJ, Kruszon-Moran D, Hill H a, McQuillan G, McAllister SK, Fosheim G, Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002J Infect Dis 2006 193:172-79. [Google Scholar]
[23]. Arakere G, Nadig S, Swedberg G, Macaden R, Amarnath SK, Raghunath D, Genotyping of methicillin-resistant Staphylococcus aureus strains from two hospitals in Bangalore, South IndiaJ Clin Microbiol 2005 43(7):3198-202. [Google Scholar]
[24]. ABCs | Bacterial Surveillance | 2008 MRSA Report | CDC [Internet]. [cited 2015 Dec 31]. Available from: http://www.cdc.gov/abcs/reports-findings/survreports/mrsa08.html [Google Scholar]