The development of extended-spectrum cephalosporins in the early 1980s was regarded as a major addition to our therapeutic armamentarium in the fight against beta-lactamase mediated bacterial resistance [1]. The emergence of enzymes that have the ability to hydrolyse these cephalosporins seriously compromised the efficacy of these life-saving antibiotics. These enzymes were called extended spectrum beta lactamases [1]. Extended spectrum beta lactamases are plasmid-mediated enzymes that are capable of conferring bacterial resistance to the penicillins, first, second and third generation cephalosporins and aztreonam [2]. They do this by hydrolysis of these antibiotics but they are inhibited in vitro by beta lactamase inhibitors [2].
Extended spectrum beta lactamases are mutant forms of broad spectrum beta lactamases such as the TEM-1, TEM-2 and SHV-1 enzymes coded by genes located on transferrable plasmids, which can easily spread from one organism to another [3]. Other ESBL enzymes described are the OXA-type, CTX-M type and PER type among others [4]. ESBLs have been reported worldwide in many different genera of Enterobacteriaceae and Pseudomonas aeruginosa [5]. However, they are most common in Klebsiella pneumoniae and Escherichia coli [6]. The prevalence varies worldwide even in closely related regions [7]. In Nigeria, an ESBL prevalence of 9.25% was recorded in a study conducted by Yusha’u et al., in Kano to screen for ESBLs production among isolates of Enterobacteriaceae [7]. In another study conducted in a tertiary health centre in Ogun state, Nigeria to determine ESBL prevalence in Escherichia coli and Klebsiella Species; an ESBL prevalence of 2.5% for Escherichia coli and 5% for Klebsiella pneumoniae were recorded [8].
Materials and Methods
Study area and sampling method
A total of 172 and 267 non-repetitive Escherichia coli and Klebsiella Species respectively were obtained from various clinical specimens during the study period from January to June, 2014. A non-probability (convenient) purposive type of sampling was used.
Study Design
The study was a hospital-based, descriptive and cross-sectional in design.
Study Area
The study was conducted in the Department of Medical Microbiology and Parasitology of the University of Maiduguri Teaching Hospital, Maiduguri (UMTH), Borno State, north-eastern, Nigeria.
Specimen Collection
The isolates were obtained from the following specimens; wound swabs, wound biopsies, aspirates, urine, cerebrospinal fluid, blood culture, sputum, ear swabs and eye swabs that were submitted for routine analysis.
Bacterial identification
The specimen were inoculated and incubated on MacConkey agar. After 24-48 hours of aerobic incubation at 36-37 °C, isolates with colonial appearance of Escherichia coli and Klebsiella Species on MacConkey agar were subjected to Gram staining reaction according to standard methods and motility testing. Suspected isolates of Escherichia coli and Klebsiella Species were confirmed by the Microbact Gram negative identification system 24ETM (Oxoid) according to the manufacturer’s instructions.
ESBL Screening and Confirmation
The cefotaxime (30μg) and ceftazidime (30μg) antibiotics were used to screen for ESBL production using the modified Kirby Bauer method. Double disk synergy test using ceftazidime (30μg), cefotaxime (30μg) and co-amoxi/clavulanate (20/10 μg) was used to confirm ESBL production. Control strains of Klebsiella pneumoniae ATCC 700603 was used as a positive control for ESBL detection while Escherichia coli ATCC 25922 was used as negative control for ESBL as recommended by the Clinical and Laboratory Standard Institute (CLSI).
DNA Extraction
DNA extraction was done using the alkaline lysis method [9].
Primer Sequence
PCR analysis for beta lactamase genes of the family TEM, SHV and CTX-M was then carried out on all phenotypically confirmed ESBL positive isolates. Primers were obtained from USA (Bioneer, Inc) and they were used for identification of TEM, SHV and CTX-M. The primer sequence is as shown in [Table/Fig-1].
Sequence of the oligonucleotide primers used for detection of extended spectrum beta lactamases genes.
| Primers | (°C) | Nucleotide Sequences (5’-3’) References | Amplicon size (bp) |
|---|
| SHV-F | 60 | CGCCTGTGTATTATCTCCCT | |
| SHV-R | 62 | CGAGTAGTCCACCAGATCCT | 293 |
| TEM-F | 60 | TTTCGTGTCGCCCTTATTCC | |
| TEM-R | 62 | ATCGTTGTCAGAAGTAAGTTGG | 403 |
| CTX-M-F | 60 | CGCTGTTGTTAGGAAGTGTG | |
| CTX-M-R | 62 | GGCTGGGTGAAGTAAGTGAC | 569 |
DNA Amplification
This was carried out in an Eppendorf thermal cycler. Amplification was carried out according to the following thermal and cycling conditions for the TEM, SHV and CTX-M gene. Initial denaturation at 94°C for 3 minutes, denaturation at 94°C for 45 seconds of 35 cycles, annealing at 60°C for 30 sec of 35 cycles, extension at 72°C for 3 minutes of 35 cycles and final extension at 72°C for 2 minutes.
Data Analysis
The information obtained from patient’s questionnaire. and the result of data from the study was entered into a computer program. Data analysis was carried out using the statistical package for social sciences (SPSSTM) version 16.0 Chicago, IL, USA, computer software. Results were presented as tables and figures where appropriate
Ethical consideration: The study protocol was reviewed and approved by the Ethical Review Committee of UMTH.
Results
A total of 439 clinical isolates of bacteria representing 172 (39.2%)isolates were identified as Escherichia coli, 178(40.5%) as Klebsiella pneumoniae, 32(7.3%) as Klebsiella oxytoca, 27(6.2%) as Klebsiella terrigena, 25(5.7%) as Klebsiella ozaenae and 5(1.1%) as Klebsiella planticola. The distribution of the organisms isolated from the various clinical specimens is as shown in [Table/Fig-2].
Distributions of the organisms isolated by specimen (N=439)
| Organisms | Swabs | Urine | Blood | CSF | Sputum | Pus | Total |
|---|
| Escherichia coli | 39 | 68 | 18 | 16 | 0 | 31 | 172 |
| Klebsiella pneumoniae | 41 | 29 | 15 | 9 | 55 | 29 | 178 |
| Klebsiella oxytoca | 9 | 9 | 7 | 2 | 4 | 1 | 32 |
| Klebsiella terrigena | 4 | 15 | 2 | 1 | 3 | 2 | 27 |
| Klebsiella ozaenae | 13 | 8 | 0 | 1 | 1 | 2 | 25 |
| Klebsiella planticola | 5 | 0 | 0 | 0 | 0 | 0 | 5 |
| Total | 111 | 129 | 42 | 29 | 63 | 65 | 439 |
CSF = Cerebro spinal fluid.
One hundred and forty seven (33.5%) clinical isolates were found positive following preliminary screening. The distribution is as shown is as follows; out of the 172 Escherichia coli; 60(35%) were found positive. However, 66(37%) of the 178 Klebsiella pneumoniae were positive, while 11(34.4%) of the 32 Klebsiella oxytoca, 5(19%) of the 27 Klebsiella terrigena, and 5 (20%) of the 25 Klebsiella ozaenae were all positive.
One hundred twenty one clinical isolates were subsequently confirmed as ESBL positive. Klebsiella pneumoniae has the highest prevalence of 59(33.1%) followed by Escherichia coli with 41(23.8%). [Table/Fig-3] summarizes the screening and confirmatory test on the various isolates.
ESBL status of Escherichia coli and Klebsiella Species following preliminary screening with ceftazidime and cefotaxime using CLSI breakpoints and confirmatory testing using the double disk synergy method (DDSM).
| Organism | ESBL Screening | ESBL Confirmation with DDSM |
|---|
| Escherichia coli (n=172) | | |
| a. ESBL Positives | 60 (34.%) | 41 (33.9%) |
| b. ESBL Negative | 112 (65.1%) | – |
| Klebsiella pneumoniae(n=178) |
| a. ESBL Positives | 66 (37.1%) | 59 (48.8%) |
| b. ESBL Negative | 112 (62.9%) | – |
| Klebsiella oxytoca (n=32) |
| a. ESBL Positives | 11 (34.4%) | 11 (9.1%) |
| b. ESBL Negative | 21 (65.6%) | |
| Klebsiella terrigena (n=27) |
| a. ESBL Positives | 5 (18.5%) | 5 (4.1%) |
| b. ESBL Negatives | 22 (81.5%) | – |
| Klebsiella ozaenae (n=25) |
| a. ESBL Positives | 5 (20.0%) | 5 (4.1%) |
| b. ESBL Negatives | 20 (80.0%) | – |
| Klebsiella planticola (n=5) |
| a. ESBL Positives | 0 (0.0%) | 0 (0%) |
| b. ESBL Negatives | 5 (100%) | – |
[Table/Fig-4] showed a picture of the double disk synergy test for Escherichia coli using cefotaxime disk, ceftazidime disk and amoxicillin/clavulanic acid in the center.
A picture of the double disk synergy test for Escherichia coli using cefotaxime disk, ceftazidime disk and amoxicillin/clavulanic acid in the center.

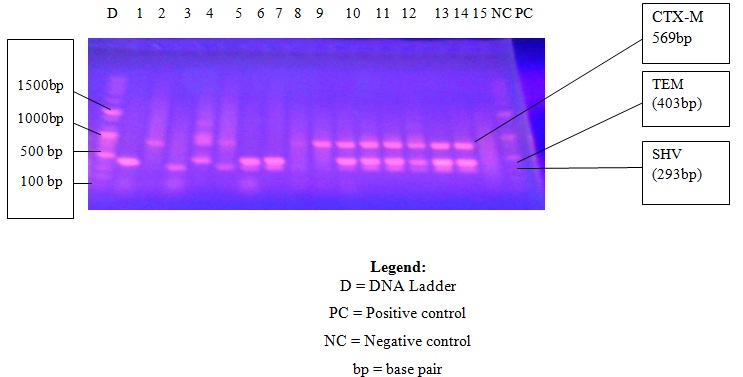
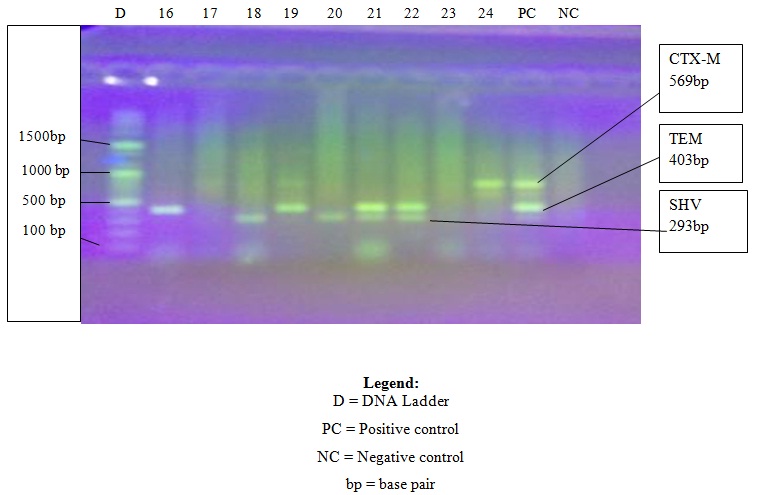
All the 121 ESBL positive isolates were subjected to molecular analysis. The result showed a highest percentage of 44(36.4%) for SHV genes followed by 38(31.4%) for TEM genes with the lowest of 33(27.3%) for CTX-M genes. [Table/Fig-5,6] shows the photograph of the PCR products for sample 1 to 23, positive and negative controls and a standard DNA Ladder.
Photograph of the PCR products showing samples 1 to 15
Photograph of PCR products showing samples 16 to 24
There were multiple occurrences of genes in some of the isolates. The co-existence of SHV, TEM and CTX-M was seen in 18 of the isolates, while CTX-M and TEM co-existed in 13 of the isolates, TEM and SHV in 7 of the isolates while CTX-M and SHV in only 3 of the isolates. Six of the isolates had none of the genes detected in them.
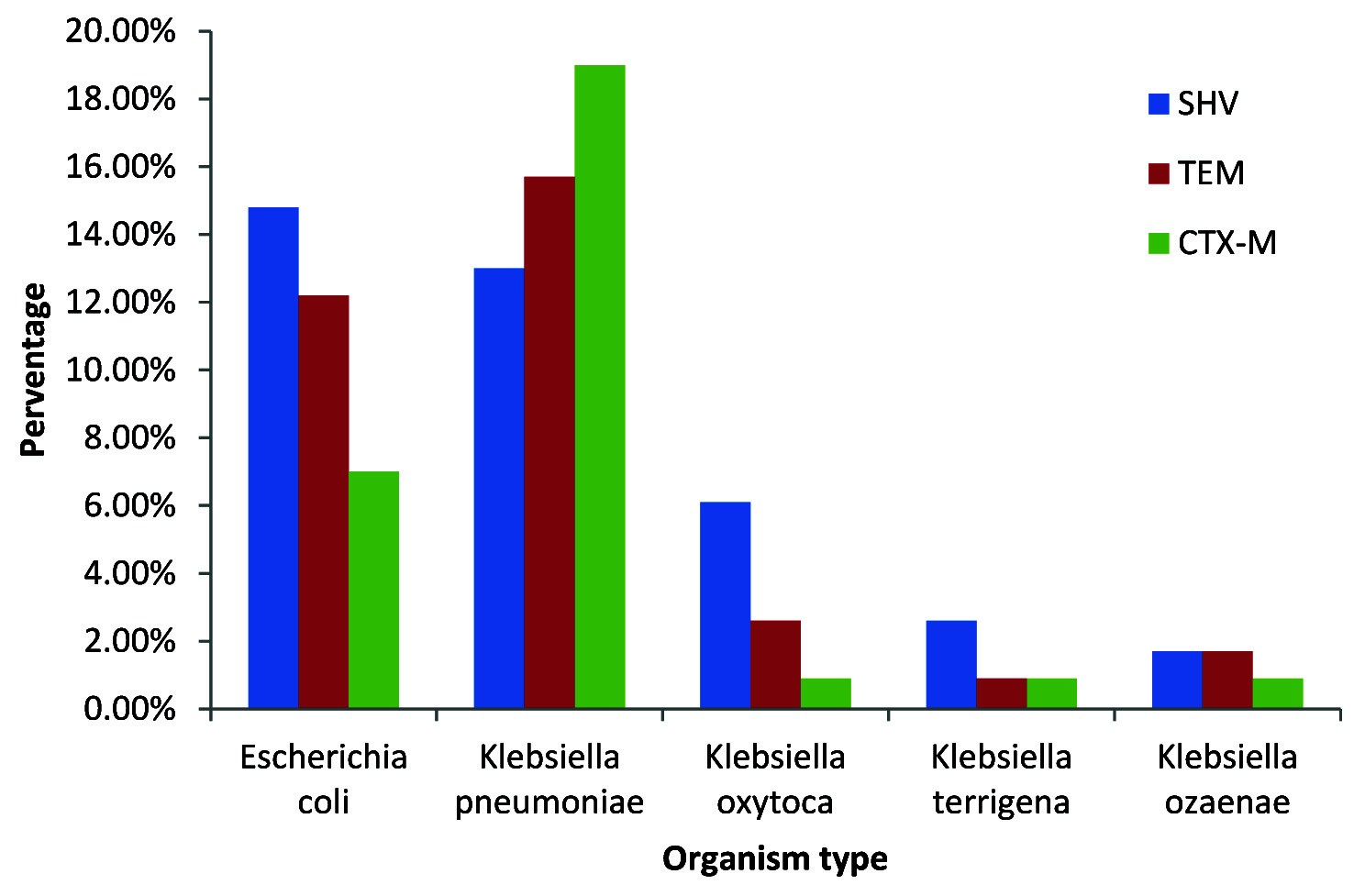
The distribution of the genes based on the organisms isolated is as shown in [Table/Fig-7]. The highest percentage of genes for Escherichia coli and Klebsiella pneumoniae were 17(38.6%) SHV for Escherichia coli and 22(66.7%) CTX-M for Klebsiella pneumoniae.
Distribution of SHV, TEM and CTX-M genes among ESBL Positive isolates of Escherichia coli and Klebsiella Species
Discussion
In the present study, a prevalence of 23.8% (41/172) and 30% (80/267) for Escherichia coli and Klebsiella Species respectively were noted. Although, it has been said that it is difficult to make valid comparison of the prevalence of ESBLs because of the variation in study design [7], the findings from this study is in agreement with the work of Olonitola [10] and colleagues. They recorded an overall ESBL prevalence of 30% (15/50) which represents 35.5% (6/17) for Escherichia coli and 27.3% (9/33) for Klebsiella Species. They used the same study design although all the isolates of Escherichia coli and Klebsiella Species were obtained from urinary specimen only. However, the prevalence recorded in this study is much higher than a prevalence of 9.25% recorded amongst genera of Enterobacteriaceae in Muhammad Abdullahi Wase Specialist Hospital, Kano [11]. Even though the study used the same method of double disk synergy that was used in this study they had a wider scope of the target organisms i.e. Enterobacteriaceae as opposed to only Escherichia coli and Klebsiella Species used in this study. The relatively high prevalence of ESBLs recorded in this study might be due to the use and misuse of third generation cephalosporins [12]. The clinical significance of the relatively high prevalence of ESBL from this study means that there would be treatment failure caused by these resistant isolates [12].
The higher ESBL rate detected in Klebsiella Species from this study is in agreement with a similar study done by Meeta and colleagues which observed the highest ESBL production in Klebsiella Species (67.04%) using similar methods of double disk synergy [13]. Although, this differs with the findings of Yusha’u in Kano, Nigeria who recorded a higher prevalence of 42.6% in Escherichia coli and 25.5% among Klebsiella Species [7]. However, the method used by Yusha’u is significantly different from the one used in this study. He utilized disk replacement methods using ceftazidime to be replaced by ceftriaxone alone and in combination with amoxicillin/clavulanic acid and he also screened for ESBL in various genera of Enterobacteriaceae as against Escherichia coli and Klebsiella Species used in this study. The reasons for high ESBL in Klebsiella Species might not be unconnected with the fact that Klebsiella pneumoniae tend to cause nosocomial infection more than Escherichia coli hence it has more chance to acquire multi drug resistance plasmids because of the hospital set up [13].
The result from this study reported a relative low occurrence of CTX-M gene, although CTX-M gene now occurs in a higher percentage in most areas of the world. A study by Vaida et al., observed the preponderance of the CTX-M type of ESBL gene amongst most isolates of Escherichia coli (96%) and Klebsiella pneumonia [14]. Several studies from Europe and Asia have also reported that CTX-M gene is now replacing TEM and SHV genes as the commonest ESBL type in that part of the world [14,15].
The explanation of our finding for lower CTX-M gene is probably due to the fact the changing pattern of CTX-M gene observed in Europe and Asia is yet to catch up with us in Nigeria. This hypothesis and the result from this study are supported by a study [16] done to determine the prevalence of extended spectrum beta lactamase production among Klebsiella isolates in some parts of South West, Nigeria where only two types of ESBLs were discovered. SHV-types ESBL were 53.0% while TEM-types were 3.0% and 44.0% did not produce ESBL. To further buttress the findings from this study, Aibunu et al., from Lagos, Nigeria analyzed eight Nigerian ESBL producing Enterobacter Species in 2001 and detected only TEM and SHV ESBLs with no CTX-M type ESBLs [17]. Another study done by Elif and colleagues in Turkey also supported the findings from this study; following phenotypic and molecular characterization of SHV, TEM and CTX-M ESBL produced by 94 clinical isolates of Escherichia coli, Acinetobacter baumanni and Klebsiella isolates [18]. ESBL was detected in 69.14% isolates using double disk synergy test, when subjected to PCR, TEM was the commonest genotype (73.43%) followed by SHV (21.87%) and CTX-M (17.18%) either alone or in combination. Even though, a case of necrotizing fasciitis was reported [19] in a Nigerian patient in the UK where Morganella morganii and Citrobacter freundii carrying the CTX-M-15 like ESBL gene were isolated from the patient. More recently in Nigeria, a case of prolonged, uncontrolled fever in an eight-year-old girl due to Klebsiella pneumoniae ESBL CTX-M -15 was described by Aboderin et al., [20]. Another hyposethis for the relative low CTX-M from this study may just mean that CTX-M is on the upward trend to not only catch up with TEM and SHV but also to replace them as in many parts of the world.
In the present study, it was observed that there were multiple occurrences of genes in some of the isolates. This finding is similar to a study by Goyal et al., where majority (57.3%) of the ESBL strains harboured 2 or more ESBL genes [21], while Bali et al., observed that about 19.2% ESBL isolates carried more than one type of beta lactamases genes. The clinical significance of the finding is that patients having organisms possessing this multiple genes are more likely to have multi drug resistance and more likely to have the propensity for widespread nosocomial transmission.
There were some of the isolates that had none of the genes tested for in them from this study. This showed that other factors such as presence of genes other than TEM, SHV and CTX-M were effective in producing resistance to beta-lactam antibiotics. A higher value of 43% for the absence of TEM, SHV and CTX-M genes were observed by Mosavia and Behnaz from Iran [22].
This variation noted in their study was due to the fact that their study area has limited type and volume of consumption of antibiotics.
Conclusion
The present study highlights a relatively higher prevalence of ESBL in Klebsiella Species followed closely by Escherichia coli following preliminary screening and confirmatory testing by disk diffusion method. The ESBL genes of SHV, TEM and CTX were detected in the majority of the phenotypically confirmed isolates of Escherichia coli and Klebsiella Species.
In view of these findings, we recommend the establishment of national guideline for the screening of ESBL. The strict compliance to antibiotic stewardship and enforcement of infection control practices should also be strengthened in all our tertiary health centers.
CSF = Cerebro spinal fluid.