Materials and Methods
Bacterial isolates
In total 81 clinical isolates were received between March 2012 and December 2013 to the Laboratory of Infectious and Tropical Diseases Research center of Ahvaz Jundishapur University of Medical Sciences, Iran, collected in different centers in Iran including Masih Daneshvari Hospital, Tehran; Iran Pasteur Institute; two private laboratories in Tehran and Tuberculosis Reference Center of Khuzestan province, southwest of Iran.
Identification of M. fortuitum
Conventional methods
The source of totally 81 clinical isolates was various samples including sputum, bronchial washing, pleural effusion, urine and wound. The following biochemical tests were performed for the clinical isolates for identification of RGM including M. fortuitum: staining for acid fast bacilli, Mycobacteria conventional phenotypic tests including growth on Lowenstein Jensen (LJ) medium (HiMedia, India) in 7 days or less, positive nitrate test, iron uptake and niacin production [17]. For quality control of the conventional tests, M.bovis BCG, M. tuberculosis H37Rv and M. kansassi (DSM44162) were used. Molecular methods can be used to identify isolates suspicious to be M. fortuitum.
Molecular methods
DNA extraction
Chromosomal DNA was extracted using the method described by pitcher et al., with slight modification to improve the susceptibility of cells to the standard digestion. In brief, after thermal inactivation, the bacterial cells were treated with lysozyme and digested with proteinase K in the presence of sodium dodecyl sulfate. The high quality DNA was purified with phenolchloroform and precipitated with isopropanol. The precipitate was washed in 70% ethanol, dehydrated, dissolved in 100 ml of Milli-Q water, and stored at -20°C until use [18].
PCR amplification using genus specific primers
Amplification process was performed by using mycobacterial genus specific primers of MSGPF, 5’-CTGGTCAAGGAAGGTCTGCG and MSGPR, 5’-GATGACACCCTCGTTGCC. These primers can amplify a 228-base-pair fragment corresponding to a sequence from the 65-KDa heat shock protein gene (hsp65) [19].
The composition of PCR mixture (50 μl) consisted of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200μM dNTPs, 10pmol of each primer, 0.5U of Taq DNA polymerase and 20ng DNA template and Millipore H2O up to 50 μl. The amplification conditions using PCR thermocycler (BIORAD, USA) were as follows: initial denaturation at 95°C for 5 min followed by 35 cycles of denaturation at 94°C for 15 sec, annealing at 58°C for 15 sec, and extension at 72°C for 30 sec and final extension at 72°C for 10 min. M. tuberculosis H37Rv, M. bovis BCG and M. kansasi (DSM44162) were used as control positive.
PCR- restriction enzyme analysis (PRA)
PRA of hsp65 was performed as described by Kim et al., with minor modifications [20]. Briefly, a 644-bp fragment of hsp65 was amplified. PCR was performed in a volume of 50 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200μM dNTPs, 10pmol of each primer, 0.5U of Taq DNA polymerase and 50ng DNA template and Millipore H2O up to 50 μl. The reaction mixture was subjected to 30 amplification cycles of denaturation at 95°C for 60 sec, annealing at 62°C for 45 sec, and extension at 72°C for 90 sec followed by a 5-min final extension at 72°C. The PCR products were digested by three restriction enzymes of Ava II, Hph I, and Hpa II (Fermentas, Canada) as previously described [20]. Digested products were electrophoresed in 3% w/v agarose gel, stained with ethidium bromide and were visualized using gel documentation system (UV Tech, UK). The pattern was compared to the restriction map algorithm constructed by Kim et al and our inhouse database as well [20]. A 100 bp DNA ladder was used as size marker (Roche, Germany).
Molecular typing of M. fortuitum isolates
RAPD-analysis
The 50ng extracted DNA from the isolates was subjected to RAPD-PCR analysis using primers 27F and 1525R as previously described [21], following the recommended optimization. The PCR products were separated by electrophoresis and photographed on gel documentation system. The major bands were identified and the previous published criterion was used for RAPD pattern interpretation [22].
ERIC-PCR
We used the amplification method using specific primers as previously described by Sechi et al., with minor modification [23]. In brief, the reaction mixture were performed in a volume of 50 μl containing 20 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200mM dNTPs, 10pmol of each primer, 0.5U of Taq DNA polymerase and 50ng DNA template and Millipore H2O up to 50 μl. The programming conditions were as initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 45s, annealing at 52°C for 1 min and extension at 70°C for 1 min, with a final extension at 72°C for 10 min. Amplified products were separated by electrophoresis in 2% w/v agarose gels after staining with ethidium bromide.
Data were analysed using Chi-square test with the help of SPSS software (SPSS Incorporation, Chicago, USA).
Clustering analyses
The DNA banding patterns of ERIC- PCR and RAPD- PCR were analysed using Bionumeric 7.5 software (Apllied maths company, Sint-Martens-Latem, Belgium) with using dice similarity index for cluster analysis and the unweighted pair group average (UPGMA) described by Hunter and Gaston [24] for clustering of M. fortuitum isolates and Pearson coefficient, UPGMA, was used for Reproducibility of each clustering analyses.
Results
The screened isolates in present study were isolated from 47 (58%) male and 34 (42%) female patients with confirmed NTM infection. The patient’s ages were from 50 to 60 years with mean of 56.4. Thirty six (44.44%) out of total isolates were conventionally identified as M. fortuitum on the basis of rapid growing in less than 7 days and standard biochemical tests for RGM. The source of M. fortuitum isolates were as follows: Broncho Alveolar Lavage {BAL} (19 isolates {52.8%}), urine (9 isolates {25%}), pleural fluid (5 isolates {13.9%}), sputum (3 isolates {8.3%}).
The presumptive M. fortuitum isolates were further analysed and verified as Mycobacteria using genus and group specific PCR targeting based on a 228-base-pair fragment from the 65-KDa heat shock protein gene.
Furthermore with the application of the PRA algorithm targeting 644 bp hsp65 DNA, all 36 isolates were clearly distinguished from other mycobacteria. All isolates had the characteristic AvaII (501, 119 bp), HphI (254, 207, 97 bp) and HpaII (270, 161, 117 bp) PRA pattern of international type strain of M. fortuitum ATCC 6841T [20].
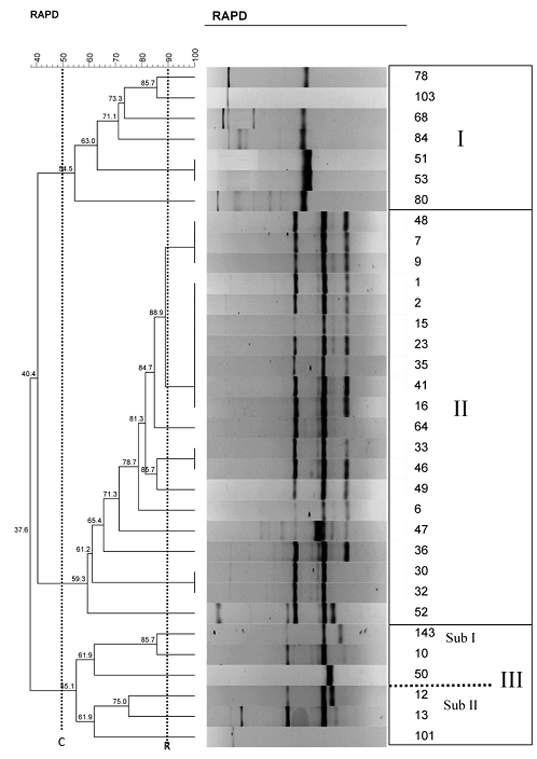
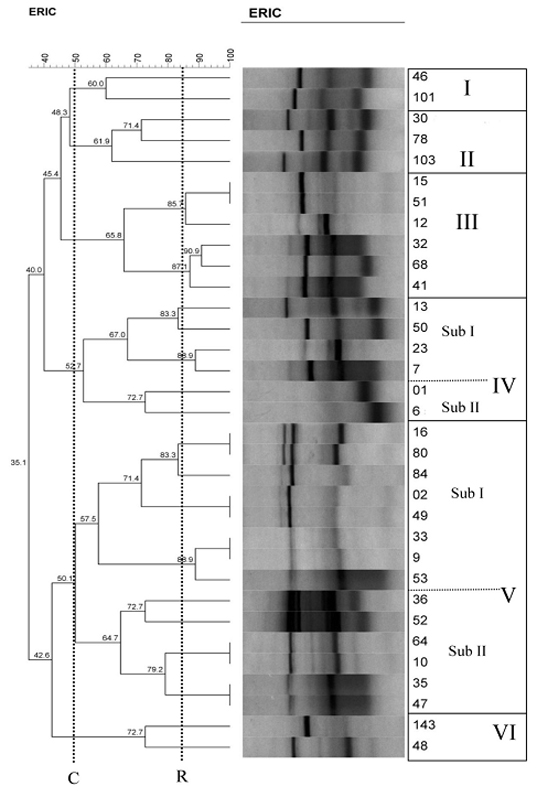
Thirty three isolates were selected for molecular typing in this study. Based on RAPD and ERIC analysis, M. fortuitum isolates were divided into 3 and 6 clusters, respectively. Most of the isolates were distributed into types of II RAPD (20 members/ 60.6%) and V (14 members/ 42.4% with sub cluster I & II) of ERIC and other types for RAPD were included: I (7 members/ 21%), III(6 members/ 18.2%, with sub cluster I & II) and for ERIC types: I (2 members/6 %), II (3 members/ 9%), III (6 members/18.2%), IV (6 members/ 18.2% with sub cluster I&II), VI (2 members/ 6%) [Table/Fig-1,2]. More than 70% of RAPD type I and II were isolated from BAL and sputum samples [Table/Fig-3]. In RAPD analysis, the major fragments were 300bp which was observed in many of RAPD types, followed by fragment 1000bp [Table/Fig-4] In ERIC analysis, the major fragments were 280bp which was observed in almost all types of ERIC-PCR, followed by fragment 1200bp [Table/Fig-5].
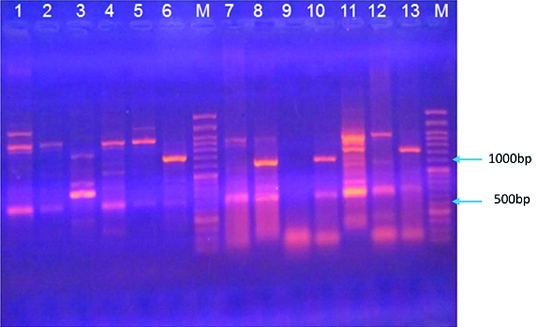
ERIC PCR profiles of Iranian Mycobacterium fortuitum isolates. Slots from left to right: 1, S80; 2, S 33; 3, S12; 4, S64; 5, S69; M, 100bp DNA size Marker; 7, S47; 8, S32; 9, S01; 10, S68; 11, S36; 12, S30; 13, S46.
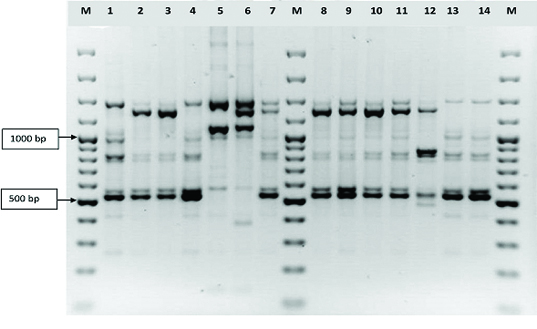
RAPD PCR profiles of the amplified DNAs of Iranian Mycobacterium fortuitum isolates obtained by analysis with primers 27F and 1525 R. Lines M, 100 bp ladder; lines 1-14, Mycobacterium fortuitum isolates pattern
Characteristics and cluster analysis of 33 Clinical isolates of Iranian M. fortuitum
| Strains | Sample source | Cluster |
|---|
| RAPD- PCR | ERIC- PCR |
|---|
| 1 | BAL | II | IV |
| 2 | Sputum | II | V |
| 6 | Sputum | II | IV |
| 7 | BAL | II | IV |
| 9 | BAL | II | V |
| 10 | BAL | III | V |
| 12 | BAL | III | III |
| 13 | Biopsy | III | IV |
| 15 | BAL | II | III |
| 16 | Blood | II | V |
| 23 | Soft tissue biopsy | II | IV |
| 30 | Sputum & BAL | II | II |
| 32 | Brain abscess | II | III |
| 33 | BAL | II | V |
| 35 | Sputum | II | V |
| 36 | Sputum & BAL | II | V |
| 41 | Blood | II | III |
| 46 | BAL | II | I |
| 47 | BAL | II | V |
| 48 | BAL | II | V |
| 49 | Sputum & BAL | II | V |
| 50 | Brain abscess | III | IV |
| 51 | BAL | I | III |
| 52 | BAL | II | V |
| 53 | Biopsy | I | V |
| 64 | Sputum | II | V |
| 68 | Sputum | I | III |
| 78 | Sputum | I | II |
| 80 | BAL | I | V |
| 84 | Brain abscess | I | V |
| 101 | BAL | III | I |
| 103 | BAL | I | II |
| 143 | Sputum | III | VI |
Abbreviations: BAL, bronchoalveolar lavage
RAPD-PCR dendrogram of Iranian Mycobacterium fortuitum isolates. The vertical dotted lines indicate the cluster cut-off value (C= shows the 50% cut-off) and the cut-off point based on the reproducibility analysis (R)
ERIC-PCR dendrogram of Iranian Mycobacterium fortuitum isolates. The vertical black (C) line shows the 50% cut-off and the cut-off point based on the reproducibility analysis (R)
Discussion
M. fortuitum group is a rapidly growing group of NTM more common in patients with genetic or acquired causes of immune deficiency [13]. For typing of Mycobacteria, several methods based on molecular biologic techniques have been introduced in recent decades including PRA, pulsed field gel electrophoresis (PFGE), RAPD and ERIC PCR. For instance, PRA schemes targeting hsp65 have been most widely used lately by some investigators with the explanation that this molecule is conserved in all mycobacteria and related strains, so it shows sufficient sequence variation to allow mycobacterial differentiation at the species or strain level [20]. In this study PRA was successfully applied to molecularly detect M. fortuitum isolates and distinguishes them from other related NTM. Although the technique could not sufficiently recognize the genetic variation among the M. fortuitum isolates and the necessity for application of another genotypic method was felt and initially the candidate was RAPD- PCR, which shows sufficient sequence variation in the strains to allow differentiation at the species or strain level. However, there was some controversy among investigators for using the method. While Zhang et al., found the technique a useful tool for typing and differentiation of species of Mycobacteria [15], it was suggested by others that RAPD-PCR alone is not sufficiently discriminative to evaluate M. fortuitum isolates from outbreaks [3]. On the other hand, some other investigators emphasized on the priority of rep-PCR on RAPD and according to their findings, rep-PCR was able to provide improved reproducibility over RAPD and a discriminatory genotyping method for mycobacterial isolates evaluated [25]. So, in present study apart from using RAPD-PCR, we have designed to apply one more widely used method i.e. ERIC-PCR to have higher discriminatory power for our M. fortuitum isolates. Other investigators reported that ERIC marker could be used as an independent marker and shows higher discriminatory power compared to other typing methods [23].
With RAPD- and ERIC PCRs combination, cluster analysis showed that ERIC was able to divide isolates into more groups and sub cluster. Results from this study demonstrated that ERIC has the potential to be used as a screening tool and useful for rapid epidemiological typing tools of M. fortuitum infections and was recommended by the authors for initial screening with many strains of M. fortuitum are to be typed. From 3 and 6 genotypic clusters generated by RAPD and ERIC PCRs respectively, the majority of isolates were belonged to types II RAPD and V, III, IV ERIC, this represents a high concordance of the results extracted from both techniques. However, considering the main and inferior fragments generated by RAPD- and ERIC-PCR, the results from ERIC was more discriminative and in general ERIC generated more uniform bands among the isolates with 2 major fragment bands of 280 & 1200 bp and at least 2 more minor fragment bands of 100 & 1000 bp. While in RAPD only 2 major fragment bands of 300 & 1000 bp were observed in almost all types and other fragment were mostly distributed in single genotypic types. We found a significant association between the type of sample and nature of generated genotype. Though the distribution of genotypes among different clinical samples showed great variation, however the isolates from BAL and sputum showed more uniformity in both genotypic techniques, since the major types isolated from these samples were type II and V, may be due to the fact that especially the number of BAL samples in this study was higher.
Although RAPD is relatively simple and useful for epidemiological analysis, standardization of the PCR conditions is very important for reproducibility as other investigators addressed [21]. Due to this, as one of the important issues observed in this work based on the experience of a previous study [22], was the necessity of standardizing the amount of DNA in each RAPD reaction mixture (50 ng of DNA) to ensure that non-specific bands were not present.
Similarity of > 50% in M. fortuitum isolates, showed high heterogeneity among them. Besides, the 100% similarity was found between some of the isolates as well, which may be due to a similar source. Our study showed that the same genetic types of M. fortuitum isolated from sputum and BAL of types II RAPD and V ERIC, occurred in different patients with 100% similarity in DNA pattern, and it was possible to postulate that the same clone was present in the environment. The results also suggest that cross-colonization was probably occurred between same patients (with 100% similarity in DNA pattern).
Conclusion
Though the results from present study represented higher discriminatory power of ERIC, however the combination of RAPD and ERIC analysis were able to sufficiently discriminate the genotypic diversity, infection control and gain useful epidemiological information regarding M. fortuitum isolates. Both techniques are simple, rapid, inexpensive, reproducible and are less complicated than most of other molecular typing methods such as PFGE or MLST. On the other hand, the cost of reagents for PFGE and MLST are expensive than the present typing techniques. Molecular epidemiology methods such as ERIC PCR, RAPD PCR and other PCR based techniques require only optimized and standard protocols and equipments available in most Institutes. Further studies of molecular characteristics of M. fortuitum are required among hospital staff and environment in the present region of the study to give us more extensive insight of actual epidemiology and state of infection-control by molecular epidemiology methods. The molecular epidemiological methods have proven to be useful for the detection and control of many outbreaks of infections by bacteria.
Abbreviations: BAL, bronchoalveolar lavage