Introduction
Type 2 diabetes mellitus (T2DM) has been recognized as a heterogeneous group of metabolic and multifactorial disorders affecting the adult population. India ranks second in the world in diabetes prevalence, just after China [1]. Genetic and environmental factors play important roles in the progression of the disease [2]. Both longitudinal and cross-sectional studies have demonstrated that T2DM is influenced by several behavioural as well as lifestyle factors [3]. Clinical and epidemiological studies have indicated that obesity is a major risk factor for T2DM, associated with an increased risk of developing insulin resistance and impaired glucose tolerance. Impaired insulin secretion and insulin resistance—the two main pathophysiological mechanisms leading to T2DM—have a significant genetic component [4].
Single nucleotide polymorphisms (SNPs) are the most common type of genetic variation among individuals of a species and are considered powerful markers for genetic mapping and genome-wide association analysis (GWAS) [5]. These polymorphisms have frequencies of >5%, accounting for approximately 90-95% of human variation [6]. Earlier GWA studies in different populations have linked many loci in the aetiology of T2DM. Genes implicated in T2DM have been found to confer moderate risk, and efforts to replicate these studies have been successful [7]. Studies have identified diabetes susceptibility loci on chromosomes 10q25.3 and 8q24.11, where transcription factor 7 like 2 (TCF7L2) and solute carrier member 30, zinc transporter, member 8 (SLC30A8) genes are located [8,9]. TCF7L2 was identified by Grant et al., [8] in an Icelandic population in T2DM subjects, and SLC30A8 was identified in the first GWAS study of T2DM in a French population [9]. TCF7L2 (rs7903146) and SLC30A8 (rs13266634) have been found to be associated mainly with impaired β-cell function [10]. Insulin growth factors are known as somatomedins, and insulin growth factor 2 (IGF2) contributes to pancreatic β-cell growth and development by regulating β-cell replication, renewal, and apoptosis [11]. IGF2 is a mitogenic peptide with important autocrine and paracrine signaling action. IGF2 ApaI, is one of the most Common polymorphism and has been analysed for its contribution to different pathologies and complications [12]. The Apa1 polymorphism was found to be associated with post transplant diabetes and rs7903146/rs13266634 polymorphisms with T2DM in a native population [13–15]. Based on the results obtained in a population studied by other group, we performed this study to analyse the association between the TCF7L2 (rs7903146), SLC30A8 (rs13266634) and IGF2 (rs680) polymorphisms with T2DM in Hyderabad, a cosmopolitan city in South India.
Materials And Methods
Patients and control subjects
This was a case-control study carried out at the Department of Genetics and Molecular Medicine, Kamineni Hospitals, Hyderabad, India. During the study period i.e. 2008-2011, we have managed to collect 500 non-obese individuals i.e. 250 T2DM patients and 250 healthy control subjects as described in our earlier publications [16,17]. Both the patients and controls were matched for sex and BMI but not for age. The diagnosis of T2DM was based on the criteria laid down by the American Diabetes Association [18]. An Endocrinologist confirmed the T2DM cases, were relevant to include in our study group. The selected cases have been diagnosed clinically with abnormal glucose values for more than 10 years with and without family history of T2DM. The selection, inclusion, and exclusion criteria for the T2DM patients have been defined in prior publications [16,17,19]. Retrospective analysis was done using the questionnaire to collect clinical, personal, and family details. Control subjects were enrolled from the general healthy population of Hyderabad; they were non-diabetic, non-obese, and free from other metabolic disorders. This study was performed with the approval of the institutional ethics committees of both the hospitals [16,17,19].
Blood sampling
The clinical and biochemical assessments of T2DM cases and control subjects have been described in detail in previous studies [16,17]. 5 mL of peripheral blood was collected from both T2DM patients and healthy controls. Among them, 3 mL serum samples were used for biochemical analyses, to confirm the disease, and remaining 2 mL blood samples with EDTA were used for the molecular analysis.
DNA genotyping
The salting out technique is the regular method used in our NABL-accredited laboratory to separate genomic DNA from the EDTA blood [20,21]. DNA was quantified using Nanodrop 2000 (Thermo Fisher Scientific, MA, USA). The genomic DNA was stored at −80°C until further processing. Genotyping was performed by polymerase chain reaction (PCR) and restriction fragment length polymorphism (RFLP) analysis followed by agarose gel electrophoresis. The 25-μL reaction mixtures (Bangalore Genei kit; Bangalore Genei Pvt. Ltd., Bangalore, Karnataka, India) were carried out with Applied Biosystems thermal cycler machine (PCR) (Life Technologies, Carlsbad, CA, USA). The primers of selected SNPs were adapted from earlier published articles [13,22] and synthesized by Bio Serve Biotechnologies (BioServe Biotechnologies, Ltd., Hyderabad, India). The details of the genes, SNPs, primers, restriction enzymes, and other components used in this study are in [Table/Fig-1]. The digested products were run on a 3% agarose gel stained with ethidium bromide and visualized under UV light.
Details of the genes and their primers involved in this study
| Gene | region | Chr Region | SNP | rs no | Forward Primer | Reverse Primer | Amplicon Size | Detection Method |
|---|
| TCF7L2 | Intron 3 | 10q25 | C>T | rs7903146 | ACAATTAGAGAGCTAAGCACTTTTTAGGTA | GTGAAGTGCCCAAGCTTCTC | 188/158 | RsaI |
| SLC30A8 | Exon 8 | 8q24.11 | C>T | rs13266634 | GAAGTTGGAGTCAGAGCAGTC | TGGCCTGTCAAATTTGGGAA | 256/176/80 | HpaII |
| IGF2 | Exon 9 | 11p15.5 | A>G | rs680 | CTTGGACTTTGAGTCAAATTGG | GGTCGTGCCAA TTACATTTCA | 292/229 | ApaI |
Statistical Analysis
Statistical analysis was carried out with SPSS software, version 21.0, for Windows (IBM Corp., New York, NY, USA). The chi-square (χ2) test was used to compare genotype frequencies in 250 T2DM patients vs. 250 controls and was used to determine whether the distribution of selected genes were in Hardy–Weinberg equilibrium (HWE) by measuring goodness of fit. Multifactor dimensionality reduction (MDR) analysis was carried out with the selected genes to determine correlation. Allele and genotype frequencies were compared between T2DM patients and controls using the Chi-square (χ2) test. A value of p<0.05 was considered to indicate a significance.
Results
Clinical characteristics
A total of 500 subjects were enrolled in this study. The demographic characteristics of risk factor variables for T2DM patients and control subjects are presented in [Table/Fig-2]. The mean age of the T2DM patients was 57.19 ± 8.22 years, with a mean body mass index (BMI) of 27.5 ± 4.1 kg/m2. The average age of the control subjects was 53.93 ± 6.32 years, with a mean BMI of 25.8 ± 3.9 kg/m2. There was a significant difference in the levels of fasting blood sugar, postprandial blood sugar, triglycerides, High-density lipoprotein (HDL)-C, and total cholesterol between the T2DM patients and controls (p < 0.05). Sex, BMI, Low-density lipoprotein (LDL)-C, and family history were not significantly associated with T2DM (p>0.05), although family history was closely associated with T2DM (58.4%).
Common characteristics of T2DM patients and control subjects
| S.No | Characteristics | T2DM Cases (n=250) | Controls (n=250) | p-value |
|---|
| 1 | Age (Years) | 41-82 (57.19±8.22) | 41-60 (53.93±6.32) | 0.0003 |
| 2 | Males/Females (%) | 55.2% / 44.8% | 57.6% / 42.4% | 0.34 |
| 3 | BMI (kg/m2) | 27.5±4.1 | 25.8±3.9 | 0.43 |
| 4 | T2DM Interval | 13.1±6.3 | NA | NA |
| 5 | FBS (mg/dL) | 143.61±55.66 | 93.54±12.13 | 0.0001 |
| 6 | PPBG (mg/dL) | 201.29±25.25 | 117.29±19.07 | 0.0001 |
| 7 | TG (mg/dL) | 156.42±78.97 | 138.77±53.69 | 0.0001 |
| 8 | TC (mg/dL) | 183.95±51.54 | 175.06±33.05 | 0.0001 |
| 9 | HDL-C (mg/dL) | 88.72±23.1 | 82.61±20.6 | 0.01 |
| 10 | LDL-C (mg/dL) | 38.76± 4.4 | 35.53±4.1 | 0.26 |
| 11 | Family History, n (%) | 58.4% | 55.2% | 0.37 |
NA= Not Analysed/ Not Applicable
Allele and genotype frequencies
The genotype distribution of TCF7L2 (rs7903146), SLC30A8 (rs13266634), and IGF2 (rs680) variants was in accordance with HWE. The genotype and allele frequencies of T2DM patients and control subjects are summarized in [Table/Fig-3] the selected SNPs (rs7903146, rs13266624, and rs680).
Allele and genotype distribution and selected polymorphisms
| Genotypes/ Alleles | T2DM cases (n=250) | Controls (n=250) | OR (95% CI) | χ2 | p-value | OR (95% CI)* | p-value* |
|---|
| TCF7L2 |
| CC | 92 (36.8%) | 144 (57.6%) | Reference | | | Reference | |
| CT | 120 (48%) | 87 (34.8%) | 2.1 (1.4, 3.1) | 15.9 | 0.0006 | 2.0 (1.4, 3.1) | 0.0006 |
| TT | 38 (15.2%) | 19 (7.6%) | 3.1 (1.7, 5.1) | 14.2 | 0.0001 | 3.0 (1.7, 5.1) | 0.0001 |
| CT+TT | 158 (63.2%) | 106 (42.4%) | 2.3 (1.6, 3.3) | 21.6 | 0.0003 | 2.1 (1.6, 3.3) | 0.0004 |
| C | 304 (61%) | 375 (75%) | Reference | | | Reference | |
| T | 196 (39%) | 125 (25%) | 1.9 (1.4, 2.5) | 23.1 | 0.005 | 1.8 (1.3, 2.6) | 0.006 |
| SLC30A8 |
| CC | 148 (59.2%) | 152 (60.8%) | Reference | | | Reference | |
| CT | 87 (34.8%) | 79 (31.6%) | 1.1 (0.7, 1.6) | 0.4 | 0.52 | 1.0 (0.7, 1.6) | 0.54 |
| TT | 15 (6%) | 19 (7.6%) | 0.8 (0.3, 1.6) | 0.3 | 0.56 | 0.9 (0.3, 1.6) | 0.57 |
| CT+TT | 102 (40.8%) | 98 (39.2%) | 1.0 (0.7, 1.5) | 0.1 | 0.71 | 0.9 (0.8, 1.6) | 0.74 |
| C | 383 (77%) | 383 (77%) | Reference | | | Reference | |
| T | 117 (23%) | 117 (23%) | 1 (0.7, 1.3) | 0 | 0.99 | 0.9 (0.7, 1.3) | 1.0 |
| IGF2 |
| AA | 135 (54%) | 166 (66.4%) | Reference | | | Reference | |
| AG | 104 (41.6%) | 78 (31.2%) | 1.6 (1.1, 2.3) | 6.8 | 0.008 | 1.5 (1.0, 2.4) | 0.009 |
| GG | 11 (4.4%) | 6 (2.4%) | 2.2 (0.8, 6.2) | 2.5 | 0.11 | 2.1 (0.8, 6.2) | 0.12 |
| AG+GG | 115 (46%) | 84 (33.6%) | 1.6 (1.1, 2.4) | 8.0 | 0.004 | 1.5 (1.0, 2.5) | 0.005 |
| A | 374 (75%) | 410 (82%) | Reference | | | Reference | |
| G | 126 (26%) | 90 (18%) | 1.5 (1.1, 2.0) | 7.6 | 0.005 | 1.4 (1.0, 2.1) | 0.006 |
* indicates adjusted for age sex and BMI
Repetition of TCF7L2 genotypes and alleles
The band sizes for TCF7L2 rs7903146 polymorphisms were 158 bp for the C allele and 188 bp for the T allele. The frequencies of CC, CT, and TT genotypes in T2DM cases were 36.8%, 48%, and 15.2%, respectively, and 57.6%, 34.8%, and 7.6% in the controls, respectively. The allele and genotype frequencies were significantly different between cases and controls. The dominant model (OR, 2.3; 95%CI, 1.6–3.3; p = 0.0003) and the TT vs. CC genotype frequencies were found to be significantly associated (OR, 3.1, 95%CI, 1.7–5.1, p = 0.0001). The T allele frequency was higher in the patients with T2DM than healthy controls (39% vs. 25%; χ2 = 23.1, OR, 1.9; 95%CI, 1.4–2.5; p = 0.005).
Amino acid substitution in Arg325Trp
The digested products for the SLC30A8 rs13266634 polymorphism produces two alleles, namely, the major C allele (176/80 bp) and the minor T allele (256 bp). There was no significant difference in the allelic or genotypic distribution of the A325T polymorphism in SLC30A8 between T2DM patients and control subjects (T vs. C: OR, 1.0; 95%CI, 0.7–1.5; p = 0.71). The genotype distribution of C325T polymorphism in SLC30A8 was assessed on the basis of sex. Genotyping analysis was carried out using the dominance model with heterozygous variants vs. CC genotypes. Female and male individuals did found to not differ in both the cases and controls [Table/Fig-4].
Differences between C325T genotypes respect to sex in cases and controls
| T2DM Patients | Males (n=138) | Females (n=112) | p-value | OR (CI=95%) | χ2 |
|---|
| CC | 79 (57.2%) | 69 (61.6%) | Reference | | |
| CT | 51 (37%) | 36 (32.1%) | 0.43 | 0.80 (0.4, 1.3) | 0.6 |
| TT | 8 (5.8%) | 7 (6.3%) | 0.99 | 1.0 (0.3, 2.9) | 0.001 |
| CT+TT | 59 (42.8%) | 43 (38.3%) | 0.48 | 0.8 (0.5, 1.3) | 0.5 |
| Controls | Males (n=144) | Females (n=106) | p-value | OR (CI=95%) | χ2 |
| CC | 92 (63.9%) | 60 (56.6%) | Reference | | |
| CT | 43 (29.9%) | 36 (34%) | 0.37 | 1.2 (0.7, 2.2) | 0.8 |
| TT | 9 (6.2%) | 10 (9.4%) | 0.27 | 1.7 (0.6, 4.4) | 1.2 |
| CT+TT | 52 (36.1%) | 46 (43.4%) | 0.24 | 1.3 (0.8, 2.2) | 1.3 |
OR, Odds ratio, 95% CI- 95% confidence interval, χ2, Chi-square analysis calculated by SPSS v.19, calculated the SLC30A8 genotypes with gender between female’s vs males as males are higher in integer
Apa 1 polymorphism
The IGF2 (Apa 1) polymorphism (A820G) genotypes and allelic frequencies for T2DM cases and controls are presented in [Table/Fig-3]. The results indicate a significant association with the dominant model (AG+GG vs. AA: OR, 1.6; 95% C, 1.1–2.4; p = 0.004), and alleles (G vs. A: OR, 1.5; 95%CI: 1.1–2.0; p = 0.005) between the T2DM patients and control subjects.
Discussion
Diabetes, a disease associated with altered glucose homeostasis, is common in India. The International Diabetes Federation estimates that around 61.3 million people in India had diabetes in 2011, and projects that by 2030, this will go up to 101.2 million [1]. T2DM is recognized as chronic persistent hyperglycaemia resulting from pancreatic dysfunction or insulin resistance, and this disease is assuming epidemic proportions [7]. T2DM is postulated to be influenced by an interplay of genetic and environmental and/or epigenetic factors, which leads to a decline in insulin action, followed by chronic pancreatic β-cell dysfunction. When the reduction in insulin function occurs, euglycaemia is maintained by increased insulin secretion [23]. The molecular mechanisms underlying the progression of diabetes remain poorly understood [24]. Different forms of diabetes such as type 1 diabetes mellitus (latent autoimmune diabetes of adults), T2DM, gestational diabetes, post transplant diabetes mellitus (PTDM), and maturity onset diabetes of the young have been ascribed to genetic defects of β-cell function or insulin resistance. However, there is no consensus on molecular diagnosis and screening to identify pre diabetic individuals and current diagnosis and screening are based on biochemical analysis [16]. In the present study, allele and genotype distributions of IGF2 in relation to T2DM in a population from Hyderabad, India, were studied for the first time. This study is akin to a previous study on TCF7L2 and SLC30A8 in a Hyderabad, south Indian population. The genotype and allele frequencies for rs7903146, rs13266634, and rs680 polymorphisms were similar to those reported in earlier studies [13–15]. The key finding was that variants in TCF7L2 (rs7903146) and IGF2 (rs680) pose an increased risk of diabetes.
Grant et al., [8] identified a strong association between the DG10S478 microsatellite marker and T2DM in Icelandic individuals. The C to T (genomic position: 114748339) substitution at SNP rs7903146 of the intron 3 (IVS3C>T) is associated with T2DM and may function through impaired glucagon-like peptide 1 secretion, which is stimulated more by fat than by carbohydrate ingestion [25,26]. TCF7L2 is present on chromosome 10q25, spanning 215.9 kb. It considered the most influential gene in determining the genetic susceptibility for T2DM today [27]. TCF7L2 is the key transcriptional factor regulating glucose metabolism through the Wnt signaling pathway and has been reported to be critical for the development of the pancreas and islets during embryonic growth [3]. Genetic variants in this gene are associated with increased risk of T2DM in a variety of study populations [28,29].
In the first published GWAS for T2DM, SLC30A8 (rs13266634) was revealed to be associated with diabetes (OR, 1.26; p = 5.0 × 10-7). SLC30A8 is a member of the zinc transporter, localized in insulin secretary cells and maps to chromosome 8q24.11. It plays a major role in transporting zinc from the cytoplasm to intracellular insulin-containing vesicles for insulin maturation, storage, and secretion from pancreatic β-cells. The rs13266634 is a non-synonymous polymorphism (C-T), substituting amino acid arginine at position 325 to tryptophan, which results in a missense R325W substitution [30,31]. TCF7L2 and SLC30A8 were studied in Hyderabad and other populations from South India [14,15,32–35]. The rs7903146 polymorphism was found to be associated with T2DM, whereas the rs13266634 polymorphism was not associated in Indian population.
The present study supports earlier studies carried out in a similar population with rs7903146 and rs13266634 polymorphisms. Two studies were carried out in north Indian populations with the rs13266634 polymorphism [34,35]. Results of these studies did not agree; Chauhan et al., found the polymorphisms to be significantly associated with T2DM in a population of 2488 individuals, while Sanghera et al., found no association in their sample size of 532 T2DM patients [34,35]. This discrepancy can be attributed to the small sample size and the low power of the latter study. However, the study by Sanghera et al., was supported by the findings of Kommoju et al., [15,35].
IGF2 is present on chromosome 11p15.5, consists of 9 exons, and represents a strong candidate polygene for cardiovascular risk. It plays a major role in regulating glucose homeostasis, the cardiovascular system, and lipid metabolism. The major function of IGF2 is growth and cell differentiation [36]. In a Caucasian population, the A allele in the IGF2 Apa 1 (rs680) polymorphism was associated with a reduction in BMI [37]. IGF2 polymorphisms have been analysed in obese children and patients with PTDM, polycystic ovarian syndrome, cardiovascular risk, weight gain, high BMI, adiposity, or obesity [13,36–39]. Our current findings are in agreement with these prior studies.
MDR is a novel and powerful statistical tool for detecting and modeling epistasis. It is a non-parametric and model-free alternative to logistic regression for detecting and characterizing non-linear interactions among discrete genetic and environmental attributes.
In this study, MDR analysis provided evidence for high-order gene–gene interactions in the absence of any statistically significance in the data. The rs13266634 polymorphism individually was not associated with genotype analysis according to MDR analysis however interaction of TCF7L2 with SLC30A8, polymorphism contributed to pathogenesis of T2DM. TCF7L2 and SLC30A8 polymorphisms together and IGF2 could be used to predict independently T2DM in the population studied. Thus, gene–gene interaction can help develop a panel of genes required to be used as T2DM disease risk marker. Graphical and radial graphic representation data are presented in [Table/Fig-5,6].
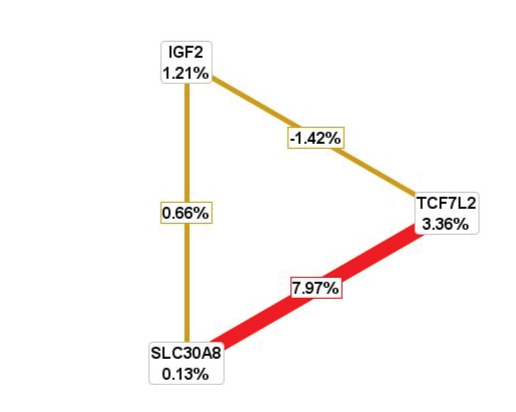
Representation of radial graphical model analysis
Legend: Combination of MDR analysis for TCF7L2, SLC30A8 and IGF2 gene polymorphism. The nodes and connections represent the entropies percentage for cases and controls enrolled in this study. Red line indicated the positive association of interaction between SLC30A8 and TCF7L2 genes and yellow color indicates poor/negative interactions in this study
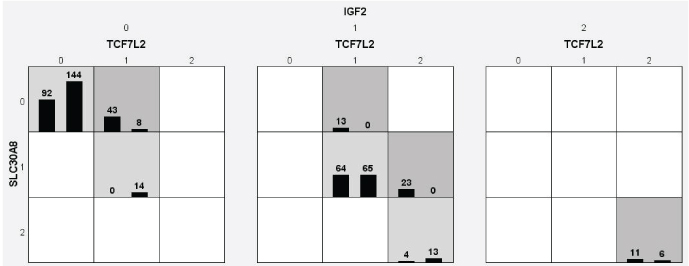
Graphical model represents the genotype distribution by MDR analysis
Legend: Square boxes representing 0 (wild type for TCF7L2 and SLC30A8), 1 (hetero for TCF7L2 and SLC30A8) and 2 (variant/mutant for TCF7L2 and SLC30A8) alleles distribution. In controls distribution of normal allele or Major allele is more common. Light shaded boxes which represents in 0-0-0 (SLC30A8, TCF7L2 and IGF2), 1-1-1 (SLC30A8, TCF7L2 and IGF2) and 2-2 (SLC30A8, TCF7L2) genotype distribution. The dark shaded boxes with 0-1-0 (SLC30A8, TCF7L2 and IGF2), 0-1-1, 1-2-1 and 2-2-2 (SLC30A8, TCF7L2 and IGF2) genotype distribution shows high risk group for the disease
Limitations
The chief limitations of the current study were that: (i) the sample size was low; (ii) only a single SNP was selected from each gene; (iii) samples were not age matched; and (iv) insulin levels were not measured.
Conclusion
To conclude, rs7903146 and rs680 polymorphisms were found independently to be significantly associated with T2DM risk in Indian adults. MDR identified the gene–gene interaction between TCF7L2 and SLC30A8 polymorphisms in confirming T2DM risk. Further studies should address the biological mechanisms affecting glucose homeostasis.
Conflict of Interest Statement
There is no conflict of interest towards this article.
[1]. Vilvanathan S, Gurusamy U, Mukta V, Das AK, Chandrasekaran A, Allele and genotype frequency of a genetic variant in ataxia telangiectasia mutated gene affecting glycemic response to metformin in South Indian populationIndian J Endocrinol Metab 2014 18:850-54. [Google Scholar]
[2]. Li YY, Gong G, Geng HY, Yang ZJ, Zhou CW, Xu J, CAPN10 SNP43 G>A gene polymorphism and type 2 diabetes mellitus in the Asian population: a meta-analysis of 9353 participantsEndocr J 2015 62:183-94. [Google Scholar]
[3]. Hussain H, Ramachandran V, Ravi S, Sajan T, Ehambaram K, Gurramkonda VB, TCF7L2 rs7903146 polymorphism and diabetic nephropathy association is not independent of type 2 diabetes-a study in a south Indian population and meta-analysisEndokrynol Pol 2014 65:298-305. [Google Scholar]
[4]. Khan IA, Jahan P, Hasan Q, Rao P, Angiotensin-converting enzyme gene insertion/deletion polymorphism studies in Asain Indian pregnant women biochemically identifies gestational diabetes mellitusJ Renin Angiotensin Aldosterone Syst 2014 15(4):566-71. [Google Scholar]
[5]. Hsu TH, Ning Y, Gwo J, AFLP-SSCP: A Useful AFLP-based method for informative SNPs discovery in non-model organismsAdvances in Biological Chemistry 2014 4:376-81. [Google Scholar]
[6]. Al-Sinani S, Hassan MO, Zadjali F, Al-Yahyaee S, Albarwani S, Rizvi S, Utility of large consanguineous family-based model for investigating the genetics of type 2 diabetes mellitusGene 2014 548:22-28. [Google Scholar]
[7]. Mayans S, Lackovic K, Lindgren P, Ruikka K, Agren A, Eliasson M, TCF7L2 polymorphisms are associated with type 2 diabetes in northern SwedenEur J Hum Genet 2007 15:342-46. [Google Scholar]
[8]. Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetesNat Genet 2006 38:320-23. [Google Scholar]
[9]. Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, A genome-wide association study identifies novel risk loci for type 2 diabetesNature 2007 445:881-85. [Google Scholar]
[10]. Kirchhoff K, Machicao F, Haupt A, Schafer SA, Tschritter O, Staiger H, Polymorphisms in the TCF7L2, CDKAL1 and SLC30A8 genes are associated with impaired proinsulin conversionDiabetologia 2008 51:597-601. [Google Scholar]
[11]. Hart LM, Fritsche A, Rietveld I, Dekker JM, Nijpels G, Machicao F, Genetic factors and insulin secretion: gene variants in the IGF genesDiabetes 2004 53:S26-30. [Google Scholar]
[12]. Toma M, Stavarachi M, Popa E, Serafinceanu C, Sonia S, Cimponeriu D, Insulin-like growth factor genetic variation, colorectal cancer and diabetesRomn Biotech Lett 2013 18:8475-80. [Google Scholar]
[13]. Vattam KK, Khan IA, Movva S, Mukkavali KK, Upendram P, Poornima S, IGF2 Apa 1 A/G Polymorphism Evaluated in ESRD Individuals as a biomarker to identify patients with new onset diabetes mellitus after renal transplant in asian indiansOpen Journal of Nephrology 2013 3:104-08. [Google Scholar]
[14]. Uma Jyothi K, Jayaraj M, Subburaj KS, Prasad KJ, Kumuda I, Lakshmi V, Association of TCF7L2 gene polymorphisms with T2DM in the population of Hyderabad, IndiaPLoS One 2013 8:e60212 [Google Scholar]
[15]. Kommoju UJ, Maruda J, Kadarkarai K, Irgam K, Kotla JP, Velaga L, No detectable association of IGF2BP2 and SLC30A8 genes with type 2 diabetes in the population of Hyderabad, IndiaMeta Gene 2013 1:15-23. [Google Scholar]
[16]. Khan IA, Vattam KK, Jahan P, Mukkavali KK, Hasan Q, Rao P, Correlation between KCNQ1 and KCNJ11 gene polymorphisms and type 2 and post-transplant diabetes mellitus in the Asian Indian populationGenes and Diseases 2015 2:276-82. [Google Scholar]
[17]. Khan IA, Movva S, Shaik NA, Chava S, Jahan P, Mukkavali KK, Investigation of Calpain 10 [rs2975760] gene polymorphism in Asian Indians with Gestational Diabetes MellitusMeta Gene 2014 2:299-306. [Google Scholar]
[18]. American Diabetes AssociationStandards of Medical Care in Diabetes-2008Diabetes Care 2008 31(1):S12-54. [Google Scholar]
[19]. Movva S, Alluri RV, Komandur S, Vattam K, Eppa K, Mukkavali KK, Relationship of angiotensin-converting enzyme gene polymorphism with nephropathy associated with Type 2 diabetes mellitus in Asian IndiansJ Diabetes Complications 2007 21:237-41. [Google Scholar]
[20]. Khan IA, Shaik NA, Pasupulati N, Chava S, Jahan P, Hasan Q, Rao P, Screening of mitochondrial mutations and insertion/deletion polymorphisms in gestational diabetes mellitus in the Asain Indian populationSaudi J Bio Sci 2015 22(3):243-48. [Google Scholar]
[21]. Khan IA, Kamineni V, Poornima S, Jahan P, Hasan Q, Rao P, Tumor necrosis factor alpha promoter polymorphism studies in pregnant womenJournal of reproductive health and medicine 2015 1:18-22. [Google Scholar]
[22]. Khan IA, Jahan P, Hasan Q, Rao P, Validation of the association of TCF7L2 and SLC30A8 gene polymorphisms with post-transplant diabetes mellitus in Asian Indian populationIntractable Rare Dis Res 2015 4(2):87-92. [Google Scholar]
[23]. Karolina DS, Armugam A, Tavintharan S, Wong MT, Lim SC, Sum CF, MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitusPLoS One 2011 6(8):e22839 [Google Scholar]
[24]. Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant riskBMC Med Genet 2008 9:59 [Google Scholar]
[25]. Gaulton KJ, Nammo T, Pasquali L, Simon JM, Giresi PG, Fogarty MP, A map of open chromatin in human pancreatic isletsNat Genet 2010 42:255-59. [Google Scholar]
[26]. Pearson ER, Translating TCF7L2: from gene to functionDiabetologia 2009 52:1227-30. [Google Scholar]
[27]. Ren Q, Xiao J, Han X, Luo Y, Yang W, Ji L, Rs290487 of TCF7L2 gene is not associated with type 2 diabetes in Chinese Han population: a case control study and meta-analysisExp Clin Endocrinol Diabetes 2013 121:526-30. [Google Scholar]
[28]. Ling Q, Dong F, Geng L, Liu Z, Xie H, Xu X, Zheng S, Impacts of TCF7L2 gene polymorphisms on the susceptibility of hepatogenous diabetes and hepatocellular carcinoma in cirrhotic patientsGene 2013 522:214-18. [Google Scholar]
[29]. Shokouhi S, Delpisheh A, Haghani K, Mahdizadeh M, Bakhtiyari S, Association of rs7903146, rs12255372, and rs290487 polymorphisms in TCF7L2 gene with type 2 diabetes in an Iranian Kurdish ethnic groupClin Lab 2014 60(8):1269-76. [Google Scholar]
[30]. Zhang X, Guan SL, Wang ZQ, You Y, Sun SL, Hui L, No association between the type 2 diabetes mellitus susceptibility gene, SLC30A8 and schizophrenia in a Chinese populationHum Psychopharmacol 2012 27:392-96. [Google Scholar]
[31]. Majithia AR, Jablonski KA, McAteer JB, Mather KJ, Goldberg RB, Kahn SE, Association of the SLC30A8 missense polymorphism R325W with proinsulin levels at baseline and after lifestyle, metformin or troglitazone intervention in the Diabetes Prevention ProgramDiabetologia 2011 54:2570-74. [Google Scholar]
[32]. Chandak GR, Janipalli CS, Bhaskar S, Kulkarni SR, Mohankrishna P, Hattersley AT, Common variants in the TCF7L2 gene are strongly associated with type 2 diabetes mellitus in the Indian populationDiabetologia 2007 50:63-67. [Google Scholar]
[33]. Bodhini D, Radha V, Dhar M, Narayani N, Mohan V, The rs12255372 (G/T) and rs7903146 (C/T) polymorphisms of the TCF7L2 gene are associated with type 2 diabetes mellitus in Asian IndiansMetabolism 2007 56:1174-78. [Google Scholar]
[34]. Chauhan G, Spurgeon CJ, Tabassum R, Bhaskar S, Kulkarni SR, Mahajan A, Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5,164 IndiansDiabetes 2010 59(8):2068-74. [Google Scholar]
[35]. Sanghera DK, Ortega L, Han S, Singh J, Ralhan SK, Wander GS, Impact of nine common type 2 diabetes risk polymorphisms in Asian Indian Sikhs: PPARG2 (Pro12Ala), IGF2BP2, TCF7L2 and FTO variants confer a significant riskBMC Med Genet 2008 9:59 [Google Scholar]
[36]. Faienza MF, Santoro N, Lauciello R, Calabro R, Giordani L, Di Salvo G, IGF2 gene variants and risk of hypertension in obese children and adolescentsPediatr Res 2010 67:340-44. [Google Scholar]
[37]. Rodriguez S, Gaunt TR, O’Dell SD, Chen XH, Gu D, Hawe E, Haplotypic analyses of the IGF2-INS-TH gene cluster in relation to cardiovascular risk traitsHum Mol Genet 2004 13:715-25. [Google Scholar]
[38]. San Millan JL, Corton M, Villuendas G, Sancho J, Peral B, Escobar-Morreale HF, Association of the polycystic ovary syndrome with genomic variants related to insulin resistance, type 2 diabetes mellitus, and obesityJ Clin Endocrinol Metab 2004 89:2640-46. [Google Scholar]
[39]. Hales CN, Barker DJ, The thrifty phenotype hypothesisBr Med Bull 2001 60:5-20. [Google Scholar]