Molecular Basis for Erythromycin Resistance in Group A Streptococcus Isolated From Skin and Soft Tissue Infections
Sunil Shivekar1, Thangam Menon2
1 PhD Scholar, Department of Microbiology, Dr. ALM Post Graduate Institute of Basic Medical Sciences, University of Madras, Taramani, Chennai, India.
2 Director and HOD, Department of Microbiology, Dr. ALM Post Graduate Institute of Basic Medical Sciences, University of Madras, Taramani, Chennai, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Sunil Shivekar, PhD Scholar, Department of Microbiology, Dr ALM PGIBMS, Taramani, University of Madras, India.
E-mail: sunisad@rediffmail.com
Background
In recent years there has been an increase in the use of erythromycin in the treatment of infections caused by bacteria other than Group A Streptococcus (GAS), which has resulted in increased resistance to this antibiotic. Erythromycin and other macrolides are alternative agents for treating GAS infections in patients, who are allergic to penicillin and its derivatives.
Aim
The main aim of this study was to identify frequency, pattern and genetic determinant of erythromycin resistance among the GAS isolated from skin and soft tissue infections.
Materials and Methods
A total 100 isolates of GAS were screened for erythromycin resistance by phenotypic and genotypic method.
Results
The results of the present study showed that 38% isolates were resistant to erythromycin. The iMLS (inducible macrolide-lincosamide-streptogramin) phenotype was predominant (55.26%) followed by M phenotype (26.32%) and cMLS (constitutive macrolide-lincosamide-streptogramin) (18.42%).
Conclusion
Phenotypic and genotypic analysis showed that the MLSB phenotype with ermB mediated mechanism of resistance was found the most common (76.31%) followed by mefA (20.51%). The ermTR genes was absent in all the isolates.
Erythromycin resistance, Genotypes, Phenotypes
Introduction
Group A Streptococcus (GAS) causes a variety of infections which are generally treated with penicillin and related β lactam antibiotics [1]. Erythromycin and macrolide are used as an alternative for patients, who are allergic to penicillin [2,3]. In recent years resistance to erythromycin has been reported in many parts of the world including India. There is a wide variation in erythromycin resistance ranging from 5%-40% has been observed in previous studies. The limitations of these studies are that they have used only phenotypic method like disc diffusion method and have not detected genetic determinants of resistance [4–8]. There are two major resistance phenotypes ie MLS B and M phenotypes and genes coding for them are erm and mef [9–11].
The main aim of this study was to assess the frequency, pattern and genetic determinant of erythromycin resistance of GAS strains isolated from skin and soft tissue infections.
Materials and Methods
Patients with skin and soft tissue infections attending the surgical OPD of a rural teaching hospital at Puducherry between January 2009 to December 2010 were included in the study. The study was approved by Institutional research and ethic committee. Patients were divided in two groups: Group I with deep cellulitis (80) cases and Group II with other superficial skin infections i.e. pyoderma, folliculitis and wound infections (20) cases. All the patients were in the age group between 31–60 year
Specimen Collection
Specimens were collected from the patients with sterile cotton swab or by fine needle aspiration inoculated on a 5% sheep blood agar and MacConkey agar plates plates. All the plates were incubated in 5% CO2 at 37oC and bacterial growth was observed at 24 hour and 48 hour. All suspected β-hemolytic streptococcal isolates were tested for sensitivity to bacitracin disc (50IU) (Hi Media Lab, India) and the latex agglutination test (Span diagnostic, India Ltd) was done to serogroup the isolates. The isolates identified as GAS were stored at -700C [12].
Erythromycin Resistant Phenotypes
All GAS strains were tested for their susceptibility to antibiotics by disc diffusion method as per CLSI guidelines. Erythromycin resistant GAS strains were tested for their resistance phenotype by double disc method. An erythromycin disc (15mg) and a clindamycin disc (2mg) (Hi Media Lab, India) were placed 16 mm apart on plates inoculated with the test strains. The plates were incubated overnight at 37°C in 5% CO2. After overnight incubation the presence of resistance to both discs was indicative of constitutive macrolide-lincosamide- streptogramin (cMLS) phenotype; susceptibility to clindamycin with no blunting of the inhibition zone around clindamycin disc was indicative of the M phenotype. Inducible macrolide-lincosamide-streptogramin phenotype (iMLS) showed resistance to erythromycin with blunting of the clindamycin zone proximal to the erythromycin disc [13].
Erythromycin Resistant Genotype
All the erythromycin resistant isolates by phenotypic method were further screened for erythromycin resistance genes mefA, ermB and ermTR by a multiplex PCR. The methods and primers used were adapted from the previous study [10]. The template DNA was extracted by alkali hydrolysis method as described earlier [14]. Each 25μl reaction mixture contained 5ul template DNA, 1ul of each ermB and mefA specific primer sets,1.5ul ermTR specific primer set, 1ul dNTP mix and 2ul Taq DNA polymerase in 5ul of 10X PCR buffer. The reactions were carried out in thermocycler (Eppendoff Germany) under the following conditions. Initial denaturation 95°C -2 minute and 30 cycles of denaturation 95oC –1 minute, annealing 55oC- 2 minute and extension 72oC –10 minute. The PCR amplicons were resolved in 1.2% agarose gel by electrophoresis. The amplicon size of ermB gene was 616bp, mefA gene was 348bp and ermTR gene was 206 bp [15].
emm Genotyping
All the isolates were subjected to emm gene PCR by using specific set of primers and product sizes ranged between 800bp to 1400bp. The emm gene PCR product was sequenced (Macrogen korea) and the first 180 bp of sequence of every strain was compared with the sequences in the CDC emm database http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm) to determine emm type [16].
Results
A total of 100 (22.83%) GAS strains were collected from a pus sample collected from 438 patients suspected of skin and soft tissue infections over a period of two years.
Erythromycin – Resistance
The result of present study showed that 38 (38%) isolates were resistant to erythromycin. The iMLS phenotype was observed to be predominant with (21/38) followed by M phenotype (10/38) and cMLS with (7/38). The iMLS and cMLS were present in 28(73.68%) of isolates. Of which 27 isolates carried ermB gene [Table/Fig-1]. None of the isolate was positive for mefA or ermTR gene. One isolate was positive for both ermB and mefA. Of the 10 M phenotypes, 7 were positive for mefA gene and ermB was present in 3 cases [Table/Fig-2].
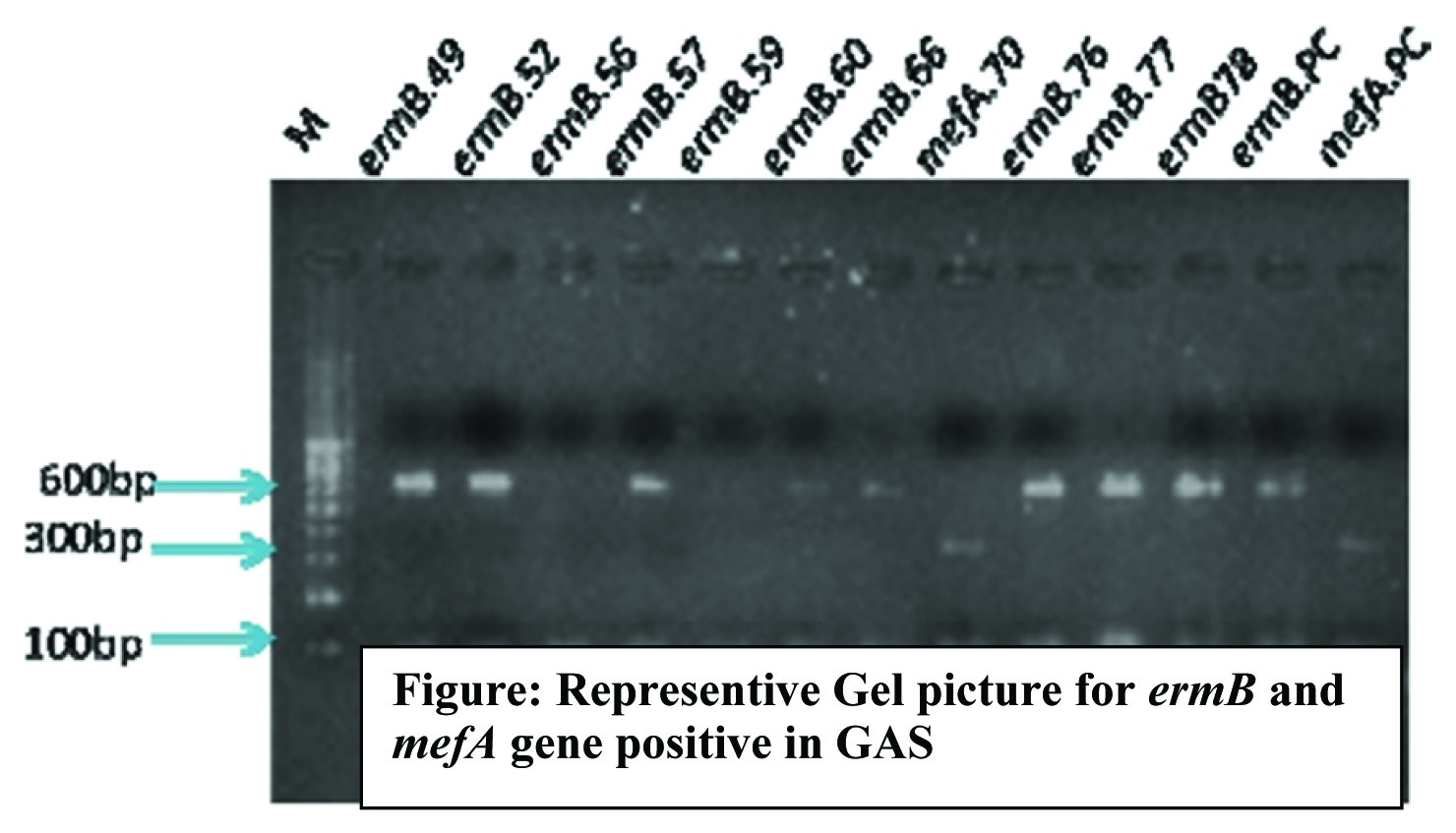
Erythromycin resistant phenotype and genotype.
| Phenotype (38) | Genotype (38) |
|---|
| mefA | ermB | mefA+ ermB | ermTR |
|---|
| M type (10) | 07 | 03 | 0 | 0 |
| iMLS (21) | 0 | 21 | 0 | 0 |
| cMLS (07) | 0 | 06 | 01 | 0 |
| Total | 07 | 30 | 01 | 0 |
The present study showed a higher rate of ermB (30/38, 76.31%) mediated genetic determinant of resistance in Indian isolates followed by mefA mediated resistance (7/30, 18.4%). The ermTR mediated mechanism was not detected in any of the isolates [Table/Fig-2].
Erythromycin Resistance and emm Types
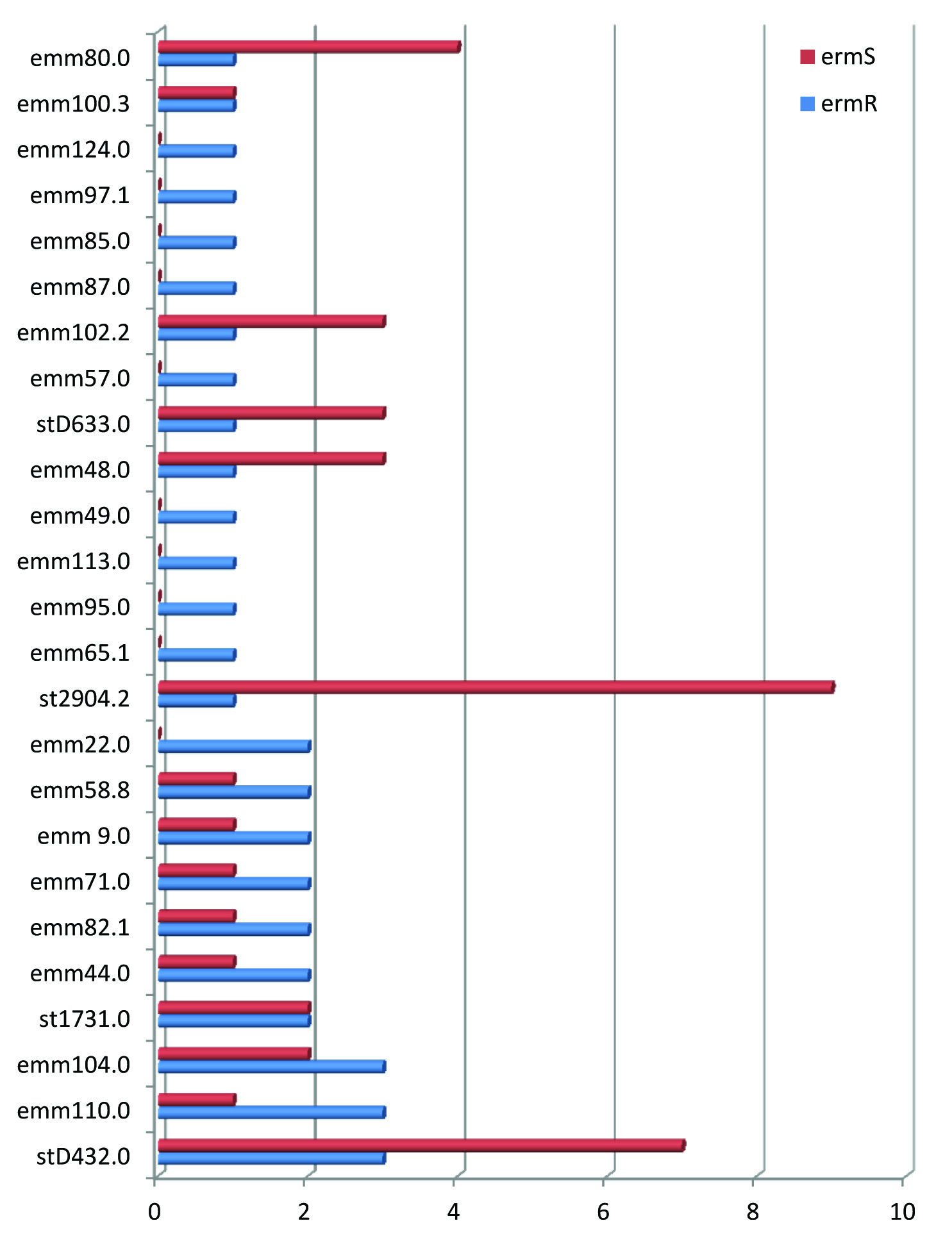
The highly heterogeneous pattern of emm types was observed with 26 different emm types. The emm 110.0, emm104.0 and st D432.0 were the most frequent and contributed for 36% of GAS showing erythromycin resistance. There was no evidence of dominance of a particular clone of emm type. Among the 26 different emm types only 6 emm types showed more than one erythromycin resistance phenotype and 20 emm types showed single erythromycin resistance phenotype indicating minimal overlapping of erm phenotypic pattern among the emm types [Table/Fig-3].
Distribuction of emm types in erythromycin resistant and sensitive strains of GAS
Discussion
Erythromycin and other macrolide antibiotic are the alternative therapeutic option to those who are allergic to penicillin. In India the use of macrolide group of antibiotics to treat pharyngeal infections has increased since last two decades [17]. The earlier reports in India on erythromycin resistance in GAS indicated a sharp increase in resistance to erythromycin from 2%-38% between 1989-2010. IIn this study we report 38% resistance of GAS to erythromycin. Similar findings were also reported in studies from Manipal (38.13%) [4] and Delhi (29.4%) [5]. The studies carried out earlier in Chennai (9.04%) [6], Vellore (13.8%) [7] and Lucknow (10.2%) [8] showed lower resistance to erythromycin compared to the present study. This is probably because all these studies were carried out on pharyngeal isolates. The present study includes isolates from skin and soft tissue infections, which are more invasive and known to have higher resistance to erythromycin. A similar resistance rate was also observed in Italy (50%), Poland (42%), Spain (27-34%) and UK (> 50%) [18,19]. The earlier studies from Chennai and Delhi has reported the predominance of M type where as present study showed the predominance of iMLS phenotype.
The previous Indian studies have not determined the genetic determinant for erythromycin resistance. This is the first study on genotypic analysis of erythromycin resistance among the GAS strains isolated from skin and soft tissue infections showing predominance of ermB gene. The similar ermB predominance was also reported earlier from Belgium, France and Italy [20–22].
The high diversity of emm types was also observed among both the erythromycin sensitive and resistance isolates. The present study also showed higher resistance rates compared to previous studies.
This may have resulted from the regular use of erythromycin in India for treating the infections other than GAS which may have caused the organisms to acquire resistance to erythromycin.
Conclusion
The result of present study indicates the high resistance of GAS against the 14- membered macrolide group of antibiotics. The infections caused by these strains can be treated with other group of macrolide such as spiramycin, josamycin and midecamycin (16-membered macrolides) with the regular surveillance for monitoring emergence of resistance.
[1]. Cunningham M, Pathogenesis of Group A Streptococcal infectionsClin Microbiol Rev 2000 13(3):470-511. [Google Scholar]
[2]. Robinson DA, Joyce AS, Wezenet T, Anand M, Debra EB, Evolution and Global Dissemination of Macrolide-Resistant Group A StreptococciAntimicrobial Agents and Chemotherapy 2006 50(9):2903-11. [Google Scholar]
[3]. Bisno AL, Gerber MA, Gwaltney JM, Kaplan EL, Schwartz RH, Diagnosis and management of group A streptococcal pharyngitis: a practice guidelineClin Infect Dis 1997 25(3):574-83. [Google Scholar]
[4]. Sharma Y, Vishwanath S, Bairy I, Biotype and antibiotic resistance pattern of group A streptococciIndian J Pathol Microbiol 2010 53(1):187-88. [Google Scholar]
[5]. Malini RC, Deepthi N, Monorama D, Kadambari B, Pushpa A, Resistance to Erythromycin and Rising Penicillin MIC in Streptococcus pyogenes in IndiaJpn J Infect Dis 2006 59(5):334-36. [Google Scholar]
[6]. Jacob SA, Lyold CAC, Menon T, cMLS and M phenotypes among Streptococcus pyogens isoalted in ChennaiIndian J of Medi Microbiol 2006 24(2):147-48. [Google Scholar]
[7]. Brahmadathan KN, Anitha P, Gladstone P, Increasing erythromycin resistance among group A streptococci causing tonsillitis in a tertiary care hospital in southern IndiaClin Microbiol Infect 2005 11(4):335-37. [Google Scholar]
[8]. Jain A, Shukla VK, Tiwari V, Kumar R, Antibiotic resistance pattern of group-a beta-hemolytic streptococci isolated from north Indian childrenIndian J Med Sci 2008 62(10):392-96. [Google Scholar]
[9]. Leclerq R, Courvalin P, Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modificationAntimicrob Agents Chemother 1991 35(7):1267-72. [Google Scholar]
[10]. Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L, Detection of erythromycin resistant determinants by PCRAntimicrob Agents Chemother 1996 40(11):2562-66. [Google Scholar]
[11]. Clancy JJ, Clancy P, Petitpas JJ, Dib-Hajj F, Yuan M, Cronan M, Molecular cloning and functional analysis of a novel macrolide-resistance determinant mefA from Streptococcus pyogenesMol Microbiol 1996 22(5):867-79. [Google Scholar]
[12]. Brooks GF, Bute JS, Morse AS, Melnick J, Adelberg’s Medical MicrobiologyAppleton and Lange publications 1998 21st edition:203-10. [Google Scholar]
[13]. Martin JM, Green M, Barbadora K, Wald A, Erythromycin-Resistant Group A Streptococci in school children in PittsburghN Engl J Med 2002 346(16):1200-06. [Google Scholar]
[14]. Hartas M, Hibble M, Sriprakash KS, Simplification of a locus-specific DNA typing method (Vir typing) for Streptococcus pyogenesJ Clin Microbiol 1998 36(5):1428-29. [Google Scholar]
[15]. Bingen E, Fitoussi F, Doit C, Cohen R, Tanna A, George R, Resistance to macrolides in Streptococcus pyogenes in France in pediatric patientsAntimicrob Agents Chemother 2000 44(6):1453-57. [Google Scholar]
[16]. Beall B, Fracklam R, Sequencing emm- specific PCR product for routine and accurate typing of Group A streptococciJ Clin Microbiol 1996 34(4):953-58. [Google Scholar]
[17]. Kotwani A, Holloway K, Trends in antibiotic use among outpatients in New Delhi, IndiaBMC Infectious Diseases 2011 11:99 [Google Scholar]
[18]. Bassetti M, Manno G, Collida A, Ferrando A, Gatti G, Ugolotti E, Erythromycin resistance in Streptococcus pyogenes in ItalyEmerg Infect Dis 2000 6(2):180-83. [Google Scholar]
[19]. Canton R, Loza E, Morosini MI, Baquero F, Antimicrobial resistance amongst isolates of Streptococcus pyogenes and Staphylococcus aureus in the PROTEKT Antimicrobial Surveillance Programme during 1999-2000J Antimicrob Chemother 2002 50(1):9-24. [Google Scholar]
[20]. Malhotra KS, Lammens C, Chapelle S, Wijdooghe M, Piessens J, Van Herck K, Macrolide- and telithromycin-resistant Streptococcus pyogenes, Belgium, 1999–2003Emerg Infect. Dis 2005 11(6):939-42. [Google Scholar]
[21]. Bingen E, Bidet P, Mihaila AL, Doit C, Forcet C, Brahimi N, Emergence of macrolide-resistant Streptococcus pyogenes strains in French childrenAntimicrob. Agents Chemother 2004 48(9):3559-62. [Google Scholar]
[22]. Zampaloni C, Cappelletti P, Prenna M, Vitali LA, Ripa S, emm gene distribution among erythromycin-resistant and -susceptible Italian isolates of Streptococcus pyogenesJ Clin Microbiol 2003 41(3):1307-10. [Google Scholar]