Prevalence of Y Chromosome Microdeletions in Idiopathic Azoospermia Cases in Central Indian Men
Prafulla Ambulkar1, Ajay Chuadhary2, Jwalant Waghmare3, Aaditya Tarnekar4, Asoke Pal5
1 Senior Research Fellow, Human Genetic Division, Department of Anatomy, Mahatma Gandhi Institute of Medical Sciences, Sevagram, Wardha, (Ms), India.
2 Professor and Head, Reproductive Biology Unit, Department of Physiology, MGIMS, Sevagram, Wardha (MS), India.
3 Professor, Department of Anatomy, Mahatma Gandhi Institute of Medical Sciences, Sevagram Wardha, (MS), India.
4 Professor, Department of Anatomy, Mahatma Gandhi Institute of Medical Sciences, Sevagram, Wardha, (MS), India.
5 Professor, Human Genetic Division, Department of Anatomy, Mahatma Gandhi Institute of Medical Sciences, Sevagram, Wardha, (Ms), India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Asoke Pal, Professor, Human Genetic Division, Department of Anatomy, Mahatma Gandhi Institute of Medical Sciences, Sevagram, Wardha-442012, (Ms), India. E-mail : asokepal@yahoo.com
Background
Genetic factor is important determinant of human male fertility, it is involved in 10-15% infertile males. Chromosome abnormalities and Y chromosome microdeletions are the main genetic causative factors for infertility. The frequency of male infertility & microdeletions in Y chromosome are also related to ethnic, geographical variations. In this study, we evaluated the prevalence of chromosomal abnormalities and microdeletions of Y chromosome in infertile azoospermia cases in central India to assess the geographical or population based variations.
Materials and Methods
We have studied 160 non-obstructive azoospermia cases to find out frequency of chromosomal abnormalities and Y chromosome microdeletions of AZF locus. G-banding method was used for exclusion of chromosomal abnormalities. One hundred and forty eight azoospermic infertile men were screened using 12 sequence-tagged-sites (STS) primers of AZFa, AZFb, AZFc region and SRY gene (Yp) region by polymerase chain reactions.
Results
Out of 160 azoospermic infertile males, 12 (7.5%) confirmed chromosomal abnormalities and Klinefelter’s syndrome was predominantly cause of azoospermia. Of the 148 infertile males, 19 (12.8%) were shown microdeletions in different AZF regions. Deletions in AZFa region were 2.02% and 3.37% was in AZFb whereas high frequencies of deletions (6.08%) in AZFc were recorded in azoospermic males. In two azoospermic males were shown microdeletions in AZFb+c loci.
Conclusion
The prevalence of Y chromosome microdeletions in azoospermic men was 12.8% in this geographical region. Klinefelter’s syndrome is important cause in male infertility. So, the screening of Y microdeletions is essential.
AZF, Infertility, Y-STS markers
Introduction
Male infertility is the major social stigma, estimated in 15% of couples having inability to conceive after 12 month despite unprotected intercourse. Out of total, 40-50% of reproductive males have abnormalities in sperm production such as reduction in quantity, motility or abnormal morphology of sperm [1]. Male infertility is associated with varicocele, hypogonadism, anorchia, cystic fibrosis, agglutination of spermatozoa, infection for mumps or disproportion levels of gonadotropic hormones {follicle stimulating hormone (FSH), leutinizing hormone (LH), testosterone}. Although, the basic reason of infertility remain unclear. Autosomal and sex chromosomal abnormalities believe 10-15% cause of male infertility [2,3]. This conformation was strongly supported by further detection and analyses of microscopic deletion in the Yq11 region of infertile men.
The genetic constitution on the Y chromosome is important for male sex determination and normal spermatogenesis. The intervening long arm of the Y chromosome are considered as the male specific Y (MSY) that controls many genes involved in spermatogenesis, also known as non-recombining region of the Y chromosome (NRY) [4]. The genes essential for spermatogenesis are located on the long arm of the Y chromosome and interval 5 and 6, region (Yq11.23) is referred to as the azoospermia factor (AZF). It has been found in many cases that similar deletions of AZF regions cause quantitative loss in spermatogenesis [5]. The AZF region was subdivided into three nonoverlapping loci-AZFa region contains ubiquitin- specific protease 9 (USP9Y) and dead box on the Y (DBY), AZFb region involves RNA binding motif on the Y (RBMY), AZFc region has the ‘deleted in azoospermia (DAZ)’ gene cluster as an important candidate gene. Proximal part of Yq11 (Yq11.21) contain AZFa locus, whereas distal Yq11 (Yq11.23) region contain AZFb and AZFc locus [6]. Most microdeletions occur de novo in distal part of AZFb and AZFc regions leads to different patterns of male infertility, from severe oligozoospermia to non-obstructive azoospermia, which are associated with spermatogenic failure [7]. Deleted in azoospermia (DAZ) family having at least 7 functional copies in interval 6D represents the most frequently deleted region in infertile men. Partial or entire deletion of the DAZ family genes is clearly associated with variable clinical phenotypes in infertile males [3,8]. However, genotype-phenotype relationship has not been fully understood. Deletions in the AZFb region have been found to be associated with azoospermia, oligozoospermia. Deletion of the AZFc region has been found to be associated with azoospermia and severe to mild oligozoospermia [9].
All over world, 8-12% incidences of idiopathic azoospermia and oligozoospermia occurs due to deletion in long arm of Yq11.23 in AZF regions. In this study, we have made an attempt to evaluate the association of Y chromosome microdeletion in azoospermia population using STS makers from each AZF region. Hence, molecular screening of Yq microdeletion is good practice to diagnose cause of spermatogenic failure in cytogenetically normal man and will help to generate epidemiological data, which in turn, will be useful for infertility clinics, who is adapted to assisted reproduction by intra-cytoplasmic sperm injection.
Materials and Methods
Our study included total 160 of central Indian origin idiopathic non-obstructive infertile men and 50 normal healthy fertile controls were included in this study. All cases visited our hospital out-patient-department and referred to reproductive biology unit for semen analysis. Their seminal examination report showed azoospermia (n=160). The age of azoospermic males ranging were 25 to 45 years. Infertile men were subjected to wide-ranging questionnaires from related to their medical, surgical, sexual and family history, lifestyle habits. Clinical exclusion criteria were strictly followed for selection of cases. Patients having chromosomal abnormalities, cystic fibrosis, testicularpathy, obstructive azoospermia, hypogonadism, hormonal disbalanced, chronic diseases were excluded from this study. Two seminal reports were considered to determine their infertility status, after 3-4 days of sexual abstinence. Seminal analysis was assessed on the basis of the guidelines of World Health Organization [10]. In case of azoospermic men scrotal sonography reports were obtained when required. Informed written consent form was obtained from each infertile male. Institutional ethical clearance committee had approved the present study.
Cytogenetics Analysis
Phytohemagglutinin (PHA) stimulated peripheral lymphocyte cultures were set up for 72 hours in RPMI 1640 medium supplemented with 10% fetal bovine serum. Chromosomal slide were prepared using standard cytogenetic technique. Ten well spread G banded metaphase were observed per individual and karyotype were prepared by microphotography [11].
Y Microdeletions analysis by polymerase chain reaction
In this study, 148 infertile azoospermic men were subjected for genomic DNA extraction by standard methods. Simplex polymerase chain reaction was done for detection of microdeletions on the AZF loci. Twelve sets of Y specific sequence tagged sites (STSs) primer were subjected to examine three AZF locus that mapped to interval 5 and 6 of the Y chromosome. The following STS primers were used: AZFa – sY746, sY84, sY86; AZFb – sY118, sY127, sY134, RBMIY; and AZFc – sY153, sY148, sY254, sY255, sY158;. Internal control was used as the SRY-sex-determining region on the short arm of the Y chromosome (sY14). All the primer sets were used from our previous studies [9]. This STS primer is recommended by the European Academy of Andrology. Over 90% of deletion in the AZF locus is making possible to detect by above primer sets and it allow for less standardization effort. A positive control of normal fertile male DNA and two negative control: (i) normal female DNA; (ii) composition of reaction mixtures except DNA were added in every PCR assay. The PCR amplification comprised a total volume of 50μL, which contained 100 to 200 ng of human genomic DNA as template, 2.5mM dNTP’s, 10 pmol/μl each forward and reverse primers (IDT, USA),10X Taq DNA polymerase assay buffer, 25mM MgCl2 and 5U Taq DNA polymerase (Merck Biosciences, India). The conditions for thermo-cycling were standardized for the each STS primer, utilizing a Veriti (Applied Biosystem, USA) thermo-cycler. Samples were subjected to polymerase chain reaction amplification using 30 cycles at 94oC for 30 sec for denaturation, 55-60oC for 45 sec for annealing and 72oC for 60 sec for aplification. Initial denaturation was done at 94oC for 5 min and final extension at 72oC for 7 min. The PCR amplified products were subjected to electrophoresis on 1% agarose gels contained 0.5μg/mL ethidium bromide and product was separated on the basis of the size. In case of detecting deletions with STS primer the PCR assay was repeated thrice for conformation.
Results
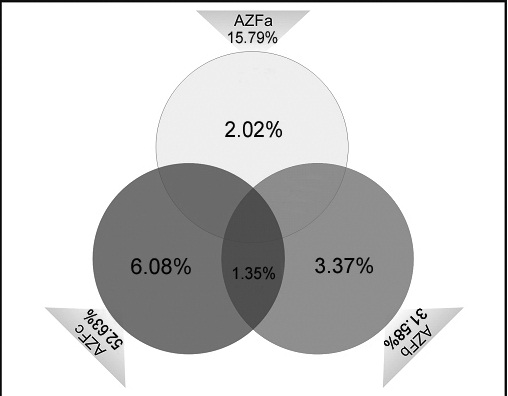
The cytogenetic abnormalities containing numerical and structural sex chromosomal aberration were observed in 12 (7.5%) individuals of 160 infertile males. However, 9 individual was 47, XXY karyotype, 1 was 46, XYq- and 2 men were 46, XY 22q+. Following the cytogenetic result, these 12 individuals were excluded from molecular studies. Polymerase chain reaction microdeletions analysis was done in 148 infertile males and 50 fertile controls. The type of cytogenetic abnormalities and AZF microdeletions are presented in [Table/Fig-1,2]. Out of 148 azoospermic men, 19 (12.8%) had microdeletions in AZFa, AZFb and AZFc regions, Three azoospermic males had microdeletions in AZFa (2.02%), here interstitial deletion was detected in sY746, sY86 STS markers in AZFa. Five azoospermic males had deletions in AZFb region (3.37%) with STS marker sY118, sY127, sY134. Nine cases had deletions in AZFc region (6.08%) with STS markers sY153, sY148, sY254, sY255 [Table/Fig.-3,4]. Deletions in both AZFb and AZFc were detected continuous in two (1.35%) azoospermic cases. No deletions were observed in the control fertile males. 12.8% (19/148) males with idiopathic non-obstructive azoospermic infertility history showed Y chromosome microdeletions with different sequence-tag-sites primers. Regionwise frequencies AZF deletions in azoospermic males were shown in Ven-diagram [Table/Fig-5]. The most frequently microdeletions (6.08%) were found within AZFc region with STS primers sY148, sY255 and sY254.
Cytogenetic finding & Karyotype for exclusion of Azoospermic men
| Types | Karyotype | Frequency | Percentage |
|---|
| Azoospermia | 47, XXY | 9/160 | 5.6% |
| Azoospermia | 46, XYq- | 1/160 | 0.62% |
| Azoospermia | 46, XY22p+ | 2/160 | 1.25% |
| Azoospermia with Normal Karyotype | 46, XY | 148/160 | 92.5% |
Regionwise microdeletions are detected in 148 azoospermia patients
| AZF region deletion | Frequency (n=148) | Percentage |
|---|
| AZFa | 3 | 2.02% |
| AZFb | 5 | 3.37% |
| AZFc | 9 | 6.08% |
| AZFb+c | 2 | 1.35% |
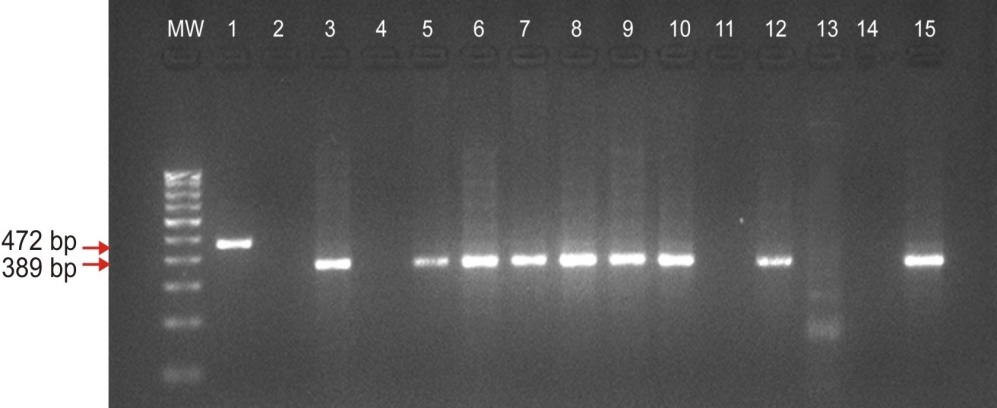
Agarose gel electrophoresis analysis shows the microdeletions in AZFc region of STS marker sY254 (389 bp) of Y chromosome in Azoospermia cases. M.W.: 100 bp of DNA ladder; Lane no 1: Internal control SRY; Lane no. 2, 4 & 11: Azoospermia with sY254 deletions; Lane 13 & 14 negative control female DNA & water sample; Lane no 15: positive control fertile male
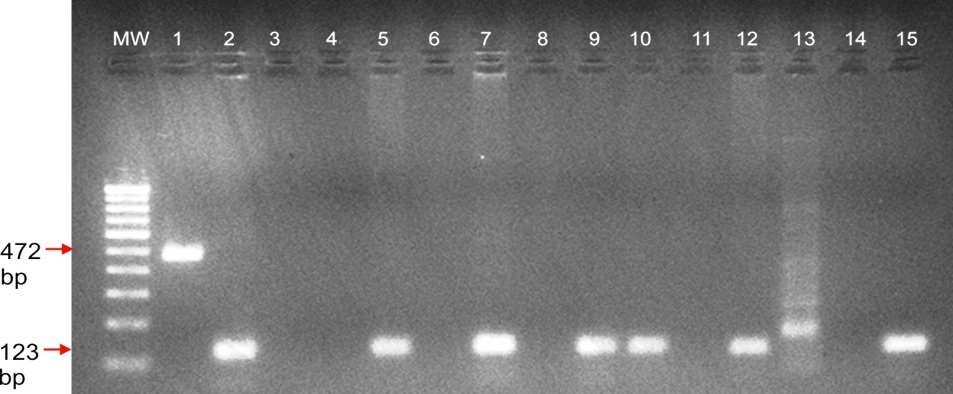
Agarose gel electrophoresis analysis shows the microdeletions in AZFc region of STS marker sY255 (123 bp) of Y chromosome in Azoospermia cases. M.W.: 100 bp of DNA ladder; Lane no 1: Internal control SRY; Lane no. 3, 4, 6, 8 & 11: Azoospermia with sY255 deletions; Lane 13 & 14 negative control female DNA & water sample; Lane no 15: positive control fertile male
Vendiagram shows percentage of azoospermic cases having microdeletions in AZFa region (2.02%), AZFb (3.37%), AZFc (6.08%) and AZFb+c (1.35%). And also the frequency distributions of microdeletions in AZFa, AZFb and AZFc regions are 15.79%, 31.58% and 52.63% respectively
Discussion
Spermatogenesis is regulated through number of genes on Y chromosome and autosomes. Microdeletions on AZF region of Y chromosome is established cause of human male infertility [12]. Spermatogenesis impairment has depended on the increasing frequency on microdeletions of Y chromosome. In male infertility, 15-30% cases involving complex genetic abnormalities can lead to different degree for spermatogenic failure [13]. By clinical findings, on the basis of semen analysis or by karyotyping Y chromosome microdeletions cannot be predicted [14]. In general, patient’s age and duration of infertility are considered important factors in making decisions regarding how to investigate, treat, and establish a reproductive prognosis [15]. Hence, in idiopathic non-obstructive azoospermic infertile male Y-chromosome microdeletions analysis has become a standard molecular test with STS markers. The prevalence of Y-chromosome microdeletions were reported in different studies, range between 10-30% of males with non-obstructive azoospermia [2,3]. Our study has evaluated the incidence of Y-chromosome microdeletions in azoospermic central Indian infertile male patients.
In present study, cytogenetic abnormalities were reported on 7.5% of azoospermia men. Nine patients were Klinefelter’s syndrome 47, XXY sex chromosomal abnormalities and in one azoospermic men had karyotype 46, XYq- which is consistent with earlier studies performed in other populations [16]. Autosomal abnormalities 46, XY (22+), found in two azoospermic patient, was already reported in azoospermic [17]. The assumption for this occurrence is that chromosomal abnormalities may interfere with normal chromosome pairing and segregation in meiosis I, leading to the potential formation of unbalanced gametes [18]. Another possibility in translocation of autosome is involved in male gametogenesis may not be regulated by breakpoint in genes [17]. The azoospermic patient with karyotype 46, XY, delY (q del) seen in the present study had lost the whole q arm with spermatogenesis involving genes. It provides the evidence for importants of Yq region for spermatogenesis and male fertility [4]. However, the relationship between chromosomal abnormalities and male infertility has been observed with a non-random distribution.
We found 19 (19/148) patients carrying microdeletions corresponding to a frequency of 12.8%. Some studies reported high frequencies of microdeletions up to more than 30% of infertile male, while others reported less than 4% in comparison to the statistical values obtained from all surveys. Another study showed an incidence of microdeletion between 5.1% and 9.6% in the infertile males. Our results are supported with these results between 3-18% [19]. In worldwide data illustrated that high frequencies of Y microdeletions are present in cases of non-obstructive azoospermia. The frequency of deletion of the AZFc region was found to be higher as compared to AZFa and AZF b region in our study, is a conformity finding with earlier studies [20]. Some earlier studies showed that deletion in the AZFc region was equal when compared with that in the AZFa and AZFb region [19]. In our present study, 3/19 (15.79%) deleted infertile males showed deletions in AZFa region, 6/19 (31.58%) deleted infertile males showed deletions in AZFb region, 10/19 (52.63%) deleted infertile males showed deletions in AZFc region and two infertile males showed deletions in both AZFb+c region. The prevalence of AZF deletion in this study population was 12.8% which is supported to previous studies of Indian population [14,21]. In another study, the prevalence of Y-chromosome microdeletions in AZF sub-regions was reported as 13.3% (12/90). A meta-analysis study has mentioned that the prevalence of Y-chromosome microdeletions was between 0.7-34.5% with an average of 8.2% [22]. However, some microdeletions are more predominant in certain populations, one has to use a more number of STS markers from each AZF region to identify Y chromosome deletions. Duplication or reduction in the copy number of DAZ genes (AZFc) was reported to have increased risk in subfertility and infertility [23].
Conclusion
However, aetiologies of a large number of azoospermic men are still unknown because of male infertility is caused by various genetic and non-genetic factors. New autosomal, X chromosomal or more Y chromosomal STS markers would help in identifying more genetic aetiologies for the idiopathic azoospermic individuals. There are variations in frequencies in different reports and these may be due to various diagnostic protocols, wrong diagnosis, selection criteria of the study population or heterogeneity in STS markers selection by various investigators. In light of the above, we also believe that the aetiology of male infertility may differ between ethnic populations. Therefore, researchers need to keep this in mind and define the strategies for analysing infertile samples. Our data will be useful for infertility clinics for genetic counselling of oligozoospermic men by advising them to choose a female child in case of Y chromosome microdeletions and to adopt appropriate methods for assisted reproduction.
[1]. Ferlin A, Arredi B, Foresta C, Genetic causes of male infertilityReprod Toxicol 2007 22:133-41. [Google Scholar]
[2]. Krausz C, Degl’Innocenti S, Y chromosome and male infertility: updateFront Biosci 2006 11:3049-61. [Google Scholar]
[3]. Sen S, Pasi AR, Dada R, Shamsi MB, Modi D, Y chromosome microdeletions in infertile men: prevalence, phenotypes and screening markers for the Indian populationJ Assist Reprod Genet 2013a 30:413-22. [Google Scholar]
[4]. Tiepolo L, Zuffardi O, Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long armHum Genet 1976 34:119-24. [Google Scholar]
[5]. Lange J, Skaletsky H, Bell GW, Page DC, MSY Breakpoint Mapper, a database of sequence-tagged sites useful in defining naturally occurring deletions in the human Y chromosomeNucleic Acids Res 2008 36:D809-14. [Google Scholar]
[6]. Thangaraj K, Gupta NJ, Pavani K, Reddy AG, Subramainan S, Singh L, Y Chromosome Deletions in Azoospermic Men in IndiaJ of Androl 2003 24(4):588-97. [Google Scholar]
[7]. Choi JM, Chung P, Veeck L, Mielnik A, Palermo GD, Schlegel PN, AZF microdeletions of the Y chromosome and in vitro fertilization outcomeFertil Steril 2004 81:337-41. [Google Scholar]
[8]. Ferlin A, Arredi B, Speltra E, Cazzadore C, Selice R, Garolla A, Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in ItalyJ Clin Endocrinol Metab 2007 92:762-70. [Google Scholar]
[9]. Ambulkar PS, Singh R, Reddy MVR, Varma PS, Gupta DO, Pal AK, Genetic Risk of Azoospermia Factor (AZF) Microdeletions in Idiopathic Cases of Azoospermia and Oligozoospermia in Central Indian PopulationJ Clin Diag Res 2014 8(3):88-1. [Google Scholar]
[10]. World Health OrganizationWHO laboratory manual for the examination and processing of human semen 2010 5th edUnited KingdomCambridge University Press [Google Scholar]
[11]. Ambulkar PS, Waghmare JE, Tarnekar AM, Shende MR, Pal AK, A Cytogenetic and Molecular Analysis of Y Chromosome Microdeletions in Idiopathic Cases of Human Male InfertilityPres Cyto & Genetics AICCG 2013 16:1-10. [Google Scholar]
[12]. Simoni M, Bakker E, Krausz C, EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions: State of the artInt J Androl 2004 27:240-49. [Google Scholar]
[13]. Massart A, Lissens W, Tournaye H, Stouffs K, Genetic causes of spermatogenic failureAsian J Androl 2012 14:40-48. [Google Scholar]
[14]. Dada R, Gupta NP, Kucheria K, Yq microdeletions—azoospermia factor candidate genes and spermatogenic arrestJ Biomol Tech 2004 15:176-83. [Google Scholar]
[15]. Kim MJ, Choi HW, Park SY, Song IO, Seo JT, Lee HS, Molecular and cytogenetic studies of 101 infertile men with microdeletions of Y chromosome in 1,306 infertile Korean menJ Assist Reprod Genet 2012 29:539-46. [Google Scholar]
[16]. Choe JH, Kim JW, Lee JS, Seo JT, Routine screening for classical azoospermia factor deletions of the Y chromosome in azoospermic patients with Klinefelter syndromeAsian J Androl 2007 9:815-20. [Google Scholar]
[17]. Rao L, Babu A, Kanakavalli M, Padmalatha V, Singh A, Singh L, Chromosomal abnormalities and Y chromosome microdeletions in infertile men with varicocele and idiopathic infertility of south indian originJ of Androl 2004 25:147-53. [Google Scholar]
[18]. Heard E, Turner J, Function of the Sex Chromosomes in Mammalian FertilityCold Spring Harb Perspect Bio 2011 3:1-17. [Google Scholar]
[19]. Zhang F, Lu C, Li Z, Xie P, Xia Y, Zhu X, Partial deletions are associated with an increased risk of complete deletion in AZFc: a new insight into the role of partial AZFc deletions in male infertilityJ Med Genet 2007 44:437-44. [Google Scholar]
[20]. Boivin J, Bunting L, Collins JA, Nygren KG, International estimates of infertility prevalence and treatment-seeking: potential need and demand for infertility medical careHum Reprod 2007 22:1506-12. [Google Scholar]
[21]. Mitra A, Dada R, Kumar R, Gupta NP, Kucheria K, Gupta SK, Y chromosome microdeletions in azoospermic patients with Klinefelter’s syndromeAsian J Androl 2006 8:81-88. [Google Scholar]
[22]. Tuttelmann F, Rajpert-De Meyts E, Nieschlag E, Simoni M, Gene polymorphisms and male infertility–a meta-analysis and literature reviewReprod Biomed Online 2007 15:643-58. [Google Scholar]
[23]. Krausz C, Chianese C, Giachini C, Guarducci E, Laface I, Forti G, The Y chromosome-linked copy number variations and male fertilityJ Endocrinol Invest 2011 34(5):376-82. [Google Scholar]