Coagulase negative Staphylococci (CoNS) are normal commensal of skin and mucous membrane and are indigenous to a variety of mammalian hosts [1]. Over the past few decades, however these organisms have gained importance as causative agents of human disease [2]. In most of the device related infections, in about 50-70% of catheter related infections CoNS are responsible as causative organisms. These infections are generally associated with the use of catheters and other medical devices [3]. The natural niche on human skin which results in easy access to prosthetic medical device inserted or implanted across the skin and adheres to biomaterials forming biofilm are the two major features owing to pathogenic success of CoNS [1].
Biofilms are sessile communities of microbial origin, characterized by the cells that are attached to a substratum in an irreversible form. They produce chemotactic particles or pheromones and the bacteria communicate with each other within the biofilm, a phenomenon called quorum sensing. They produce a matrix of extracellular polymeric substances (EPS) called polysaccharide intercellular adhesin (PIA) [4]. PIA is synthesized from UDP-N-acetylglucosamine by N-acetylglucosaminyltransferase which is encoded by ica gene [5].
The biofilm act as a barrier for eradication of bacterial cells and confers high level of resistance to antimicrobial agents. So the microorganisms growing in a biofilm exihibits intrinsically more resistant to antimicrobial agents than planktonic cells. Bacteria in biofilm grows slowly and slower growth may lead to decreased uptake of the drug and other physiologic changes that could affect drug resistance [1].
The present study was undertaken to detect the prevalence of biofilm producing and nonproducing CoNS which were isolated from various clinical samples collected from the patients admitted in various wards at Government Rajaji Hospital, Madurai and to compare the various phenotypic methods in the identification of biofilm producing CoNS with PCR.
Materials and Methods
Collection of Samples: After obtaining ethical clearance, a total of 456 clinical samples were collected from inpatients admitted at Government. Rajaji Hospital during the study period from June 2011 to May 2012. The samples include pus (from infected bone and joint prosthetic implants, surgical site infections), indwelling catheter, blood, urine.
Bacterial isolates: Of the 456 clinical samples, a total of 252 non repetitive clinical strains of Staphylococci were isolated, of which 96 were identified as Coagulase negative staphylococci [9]. The isolates from samples were identified and characterized by using standard microbiological procedures (Gram staining, catalase, coagulase, DNase and based on biochemical tests such as production of phospahatase, susceptibility to Novobiocin, ornithine decarboxylase test and beta hemolysis on blood agar, different species of CoNS were identified [9]. The CoNS isolates were subjected to antibiotic susceptibility test by Kirby Bauer’s disc diffusion method on Mueller Hinton (MH) agar and the zones were interpreted as per CLSI guidelines [10]. The following antibiotic discs were used: Ampicillin (AMP 10μg), Chloramphenicol (C30 μg), Gentamicin (GM 10μg), Ciprofloxacin (CIP 5μg), Cefotaxime (CTX 30μg), Cotrimoxazole (COT 25μg) Erythromycin (ERM 15 μg), Doxycycline (DOX 30μg) and Staphylococcus aureus (ATCC 25923) used as control strain.
Detection of biofilm production
1. Phenotypic methods
A)Congo red agar method [11]: A simple qualitative method for detection of biofilm production was described by Freeman et al., [11] and medium used was Congo Red Agar (CRA) which composes of Brain heart infusion broth (37 gms/L), sucrose (50 gms/L), agar no.1 (10 gms/L) and congo red stain (0.8 gms/L, Himedia). Biofilm producers form black colonies with a dry crystalline consistency and non biofilm producers form pink coloured colonies. The control strains were included in the test {Staphylococcus epidermidis ATCC 35984 (biofilm producer) and ATCC 12228 (biofilm nonproducer)}.
B) Tube method [12]: Christensen et al., [12] described a qualitative method for detection of biofilm production. The test organisms were inoculated in trypticase soy broth and kept for overnight incubation. Then the tubes were decanted and washed using phosphate buffer saline (pH 7.3). Tubes were stained by using safranin (0.1%) and excess stain were removed. Tubes were kept in inverted position. The control strains were included in the test and according to the results the scoring was done. Biofilm formation was considered positive if the wall and the bottom of the tube were lined by a visible film. The amount of biofilm formed was scored as 1- weak/none, 2- moderate, 3-strong.
C) Tissue culture plate method [13]: This is a quantitative method for biofilm detection [13]. Overnight growth of test organisms in trypticase soy broths was diluted 1in 100 and 200μl were inoculated in sterile 96 well flat bottom polystyrene tissue culture (Erose) and then incubated. Then the organisms used for control (positive and negative) were diluted and added to the microtitre plate and kept for incubation. The contents of the well were was removed and washed 4 times with 0.2 mL of phosphate buffer saline (pH 7.2). Sodium acetate (2%) was used to fix adherent bacteria in the wells and allowed to dry. Crystal violet (0.1%) was used to stain the wells. Excess stain was removed and air dried. Reading was taken at wavelength 570 nm by micro ELISA autoreader (LISA). As the bacteria forms biofilm and adheres to the wells, these OD values were taken as an index of bacterial adherence to the wells.
Interpretation:
Mean OD values Biofilm production
<0.120 Non/weak
0.120–0.240 Moderate
>0.240 High
2) Genotypic method:
A) Polymerase chain reaction (PCR) [14,15]: DNA was extracted by using DNA purification kit (PureFast® Bacterial Genomic DNA purification kit). PCR was done to detect ica gene in CoNS isolates. It includes the following steps: DNA extraction, PCR amplification and gel electrophoresis to visualize the amplified products. The primers used were iCA-TCCAGAAACATTGGGAGGTC (forward), ica-TGGGTATTCCCTCTGTCTGG (reverse). These primers specific for the icaADBC region were used in PCR to produce amplicon of size 516bp. The PCR cycling conditions used were 30 cycles of 1 min of denaturation at 94°C, annealing for 1 min at 58°C followed by 1 min extension at 72°C with final extension for 5 min at 72°C. PCR was performed by using thermal cycler with Taq polymerase (Helini) and the reaction mixtures were analyzed by 1% agarose gel electrophoresis. The test was done in triplicates and the control strains (positive and negative controls) were included.
Results
Out of 96 CoNS isolates, 76 isolates (79.17%) were Staphylococcus epidermidis followed by Staphylococcus haemolyticus, Staphylo-coccus lugdunensis, Staphylococcus saprophyticus as shown in [Table/Fig-1].
Sample wise identification of CoNS species n=96
| Sample | S. epidermidis | S. haemolyticus | S. Lugdunensis | S. Saprophyticus | Others | Total |
|---|
| Pus | 38(39.58%) | 3(3.125%) | 2(2.08%) | - | 6(6.25%) | 49 |
| Blood | 22(22.92%) | 2(2.08%) | 1 (1.04%) | - | 3(3.13%) | 28 |
| Urine | 16(16.67%) | - | - | 3(3.13%) | - | 19 |
| Total | 76(79.17%) | 5(5.21%) | 3(3.13%) | 3(3.13%) | 9 (9.38%) | 96 |
Detection of biofilm production by phenotypic methods: Tissue culture plate method detected biofilm production in 38(39.57%) isolates, tube method detected 29(30.2%) isolates and congo red agar method detected 17 (17.7%) biofilm producing CoNS isolates [Table/Fig-2].
Comparison of biofilm production in CoNS isolates by different phenotypic methods n=96
| Biofilm production | Tissue Culture plate (TCP) | Tube method (TM) | Congo red agar(CRA) |
|---|
| No. of isolates (96) | Positive | 38(39.57%) | 29(30.2%) | 17(17.7%) |
| Negative | 58(60.42%) | 67(69.79%) | 79(82.29%) |
Biofilm producing CoNS were isolated from pus18 (18.75%) followed by blood 11 (11.46%), urine9 (9.37%). Biofilm production was highest in Staphylococcus epidermidis 37(38.54%) followed by Staphylococcus saprophyticus1(1.04%) as shown in the [Table/Fig-3].
Sample and species wise isolation of biofilm producing CoNS n=96
| Specimen | S.epidermidis | S.saprophyticus | Percentage |
|---|
| Pus | 18 | - | 18.75% |
| Blood | 11 | - | 11.46% |
| Urine | 8 | 1 | 9.37% |
| Total | 37 | 1 | 39.58% |
Biofilm producing isolates were associated with risk factors such as infected orthopaedic implants (39.47%), urinary catheterization (21.05%) and prosthetic valve implantation (15.79%) [Table/Fig-4].
Detection of ica gene: PCR was done to identify ica gene in CoNS isolates which were found to be biofilm producers by phenotypic methods. Among the 35 PCR positive Biofilm producing CoNS isolates, TM identified 6 isolates, CRA 18 isolates as false negative and none of the isolate was identified as false negative by TCP. Among the PCR negative isolates, TCP method identified 3 isolates as false positive. The false positive identified by TM and CRA were 5 and 7 respectively.
Comparision of risk factors with biofilm producing CoNS isolates n =38
| Risk factors | No. of Biofilm producer | Staphylococcus epidermidis | Staphylococcus saprophyticus. |
|---|
| Infected Orthopaedic implants | 15(39.47%) | 15(39.47%) | - |
| Urinary catheterization | 8(21.05%) | 7(18.42%) | 1(2.63%) |
| Prosthetic valve | 6(15.79%) | 6(15.79%) | - |
Statistical Analysis
The TCP method had the highest sensitivity and specificity, the PPV and NPV were 92.1% and 100% respectively. TM showed 80% sensitivity and 92.42% specificity with 82.75% PPV and 97.04% NPV. CRA method had the least sensitivity (35.71%) and specificity (89.71%) with PPV and NPV of 58.82% and 77.22% [Table/Fig-5].
Sensitivity and specificity of various phenotypic methods
| Methods | Sensitivity | Specificity | PPV | NPV |
|---|
| TCP | 100% | 95.08% | 92.1% | 100% |
| TM | 80% | 92.42% | 82.75% | 97.04% |
| CRA | 35.71% | 89.71% | 58.82% | 77.22% |
Antibiotic susceptibility test: Of the 96 CoNS isolates, 43 (44.79%) isolates were found to be multidrug resistant. Higher antibiotic resistance was noted in Staphylococcus epidermidis (86.04%) followed by and other species of CoNS [Table/Fig-6].
Species wise isolation of multidrug resistant n=43
| CoNS Species | No. of multidrug resistant isolates |
|---|
| Staphylococcus epidermidis | 37(86.04%) |
| Staphylococcus saprophyticus | 1(2.33%) |
| other CoNS | 5(11.63%) |
Out of the 43 multidrug resistant CoNS species, 31 isolates were found to be biofilm producers, 8 isolates were non biofilm producer. Biofilm producing isolates showed higher antibiotic resistance (72.1%) than non biofilm producing isolates (18.6%). The ica gene was present in all the CoNS isolates were found to be multidrug resistant [Table/Fig-7].
Percentage of multidrug resistant biofilm producing CoNS isolates
No. of multidrug resistant strains with biofilm production n =43
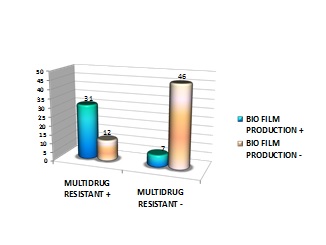 |
| Biofilm production | Multidrug resistant | Multidrug resistant |
|---|
| + | - |
| + | 31 | 7 |
| - | 12 | 46 |
Discussion
Currently, CoNS are predominant cause of nosocomial bacteremia and infect a wide variety of prosthetic medical devices [1]. The major virulence factor is the ability to adhere on the surface of biomaterials and form biofilms [16]. In this study we evaluated the presence of ica gene in comparison with different phenotypic methods for the detection of biofilm.
In this study, Staphylococcus epidermidis was the most commonly isolated species 76 (79.17%) and isolated from joint and wound infections 38 (39.58%). The second most common isolate was S.hemolyticus followed by S.lugdunensis and S.saprophyticus. This was in concordance with the studies conducted by Mohan et al., and Marsik et al., [17,18].
The present study showed that 38 (39.58%) of CoNS isolates were biofilm producers. This finding correlated with studies conducted by Seetha et al., [19], Mohan et al., [17], Makhija SK et al., [20] who showed that 43(42.5%) CoNS isolates were slime producers.
In the present study, the percentage of biofilm production detected by Tissue culture plate method (39.58%) was high followed by Tube method (30.21%) and Congo red agar method (17.71%). This finding correlated with Mathur et al., [21] showed that the number of biofilm producers identified by TCP method was high (53.9%) and followed by Tube method (11.8%) and CRA (5.17%). In another study, conducted by Ruzicka et al., [22] noted that out of 147 isolates of S. epidermidis, TM detected biofilm formation in 79 (53.7%) and CRA detected in 64 (43.5%) isolates and reported that TM is better for biofilm detection than CRA. Similar findings were also noted by Oliveira et al., of the 100 CoNS isolates 82% were biofilm positive by PCR and the tube test, 81% by the TCP assay, and 73% by the CRA method [23].
In this study, Staphylococcus epidermidis 37 (38.54%) was the most common biofilm producing CoNS species followed by Staphylococcus saprophyticus 1 (1.04%). Biofilm production had a strong association with medical device related infections such as infected implants, urinary catheterization and prosthetic valve. This in correlation with study by Sujata et al., [24] Who had documented that out of 55 S.epidermidis isolates from various device related infections, 26 (65.2%) were biofilm producers.
In the present study 43 (44.79%) CoNS isolates were found to be multidrug resistant and Staphylococcus epidermidis 37 (38.54%) was the most common multidrug resistant CoNS species. It was observed that there was higher antibiotic resistance in biofilm producing CoNS isolates than non-Biofilm producers. Similar findings were also noted in the study conducted by Sujata et al., [24], Afreenish Hassan et al., [25]. Of 43 (44.79%) multidrug resistant CoNS isolates 31 isolates were biofilm producers and showerd ica gene positivity. This is in correlation with the study conducted by Sharma et al., [26] who had noted that more than 80% invasive CoNS strains which had ica and mecA genes were resistant to multiple antibiotics and were positive for biofilm formation.
In this study, out of 38 Biofilm producing CoNS isolates detected by phenotypic method, ica gene was identified by PCR in 35 (36.45%) isolates and The results were concordance with the study by Sujata et al., [24] who had reported in their study that ica gene was present in 23 (41.8%) among 26 (47.2%) of biofilm producers. The results were in discordance with the study by Seung- Hak- Cho et al., [27] who had reported that 18 S. epidermidis isolates obtained from catheter-related urinary tract infections showed the ica speficic DNA and only 11 isolates biofilms spontaneously under normal growth conditions. But in a study conducted by Agarwal et al., [28] showed that the use of glucose or NaCl or combination of both enhanced biofilm producing capacity of staphylococcal isolates irrespective of presence or absence of ica operon.
Galdbart JO et al., [5] in their study showed that 44 out of 54 S. epidermidis isolates from prosthetic material related joint infection showed ica positivity. Gad et al., [29] Cafiso et al., [30] demonstrated the detection of ica operon by molecular technique (PCR) with high efficiency. In addition, these genes are important virulence markers of clinical CoNS isolate since their expression is associated with the production of PIA, the most clearly characterized component of staphylococcal biofilms.
Conclusion
PCR detects ica genes, the virulence marker of staphylococcal infection and biofilm non-producers are negative for icaA and icaD and lack the entire ica ADBC operon. Although the genotypic methods will be absolute detection methods, it was not done in all centres. Our present study showed that ica gene was present in 35 (36.45%) of CoNS isolates which were detected as biofilm producers by Tissue culture plate and sensitivity and specificity of biofilm detection by this method was high in comparison with Tube method and Congo red agar.
TCP can be recommended as a general screening test for biofilm production than CRA and TM. Considering the cost and specialized man power and sophisticated infrastructure, TCP can be performed to detect the Biofilm producing strains of CoNS with same sensitivity and specificity coinciding with genotypic methods.