Background
Haemophilia A (HA) is the most severe sex-linked bleeding disorder that is characterized with non-controlled and often threatening Haemorrhage. Routine fetal sex determination in early pregnancy with Haemophilia is based on invasive procedures that can be dangerous to the mother and fetus.
Aim
The goal of this study is to present an improved assay for the non-invasive fetal sex determination using a Real-Time duplex PCR on the free fetal DNA (ffDNA) obtained from the maternal serum of the HA carriers.
Materials and Methods
Blood samples were eventually collected from 23 pregnant HA carriers between the 8th and 12th weeks of gestation, and after amplification by duplex-PCR of the single copy of Y chromosome-specific sequence (SRY), the product was then subjected to Real-Time PCR analysis.
Results
Data were compared with the outcome of chorionic villus sampling (CVS) and indicated that the SRY sequence was detected in 6 of 6 serum samples from male pregnancies and that sequence was absent in 9 samples where the fetus was female. The remaining samples determined without having the CVS positive samples.
Conclusion
We tried to develop a Real-Time duplex PCR for accurate diagnosis of fetal gender early in the pregnancy of HA carriers. This study has brought up two remarkable points, the first is the method’s improvement with high specificity in sex determination, especially in screening of prenatal sex-linked disorders in male gender and the second is that fresh serum samples would be a good source for this purpose, advocated by similar studies carried out in this regard.
Cell-free fetal DNA, Congenital bleeding disorders, Maternal serum
Introduction
Haemophilia A is one of the most prevalent congenital bleeding disorders caused by a deficiency in coagulation factor VIII. Due to its X-linked inheritance, it affects approximately 1 in 5000 males, while females are typically asymptomatic carriers of the disorder [1]. The reported Haemophilia A prevalence (per 100 000 males) in the literature varies considerably among countries and according to the global survey presented by World Federation of Haemophilia about Haemophilia cases, Iran was ranked second (next to Egypt) in the eastern Mediterranean region [2,3]. Fetal sex determination is an important step in the prenatal diagnosis of Haemophilia. If the fetus is female, further investigation is not required, but in the case of a male fetus, subsequent DNA analysis is required, and the parents must be appropriately counseled. Currently however, prenatal diagnosis in the first-trimester of gestation is based on invasive procedures that carry a risk of approximately 1% for miscarriage [4]. The discovery of free fetal cells in 1950s, and free fetal DNA (ffDNA) in the 1990s, in the maternal circulation early in gestation, has opened up new possibilities for prenatal diagnosis. Although the use of fetal cells for prenatal diagnosis has necessarily been limited due to their rarity in the maternal circulation; their persistence during subsequent pregnancies; their apoptotic state, and their requirement for special isolation and enrichment procedures, the use of ffDNA for prenatal diagnosis of genetic diseases is preferable and is now well-established [5–7]. Fetal DNA originates mainly from apoptotic placental cells (trophoblasts) and circulates in maternal plasma predominantly as fragments of lengths less than 313 bp [8,9]. Probably, they are protected within apoptotic bodies or are attached to nucleosomes, so that fetal nucleic acids are unexpectedly stable in the maternal circulation [10]. Fetal DNA can be reliably detected in the maternal circulation from the 5th week of pregnancy, and its concentration increases in the course of gestation [11]. Most studies have used Y chromosome-specific fetal gender [12–15]. Initial studies using conventional PCR techniques were less sensitive and specific relative to more recent techniques, such as Real-Time PCR. Fragmentation of the ffDNA and its low concentration (3-6%) against a high background of maternal free DNA (94–97%), are the main problems involved in fetal sex determination [16,17]. To address these problems, numerous studies have so far shown that Real-Time PCR increases the specificity and sensitivity of non-invasive fetal sex determination, but for getting more specificity we tried to improve the method using Real-Time duplex PCR with general SYBR-Green assay for detecting the SRY gene in plasma and serum of the pregnant HA carriers between the 8th and 12th weeks of gestation, the period of time in which external genital development occurs and is the most favourable time of chorionic villus sampling for prenatal diagnosis of fetal sex.
Materials and Methods
Collection and processing of blood samples. A total of 23 blood samples were collected over one year (started in 2012) from the pregnant Haemophilia-A carrier, with their consent, referred to the Iranian Comprehensive Haemophilia Care Center (ICHCC) at between 8th and 12th weeks of gestation. All pregnant had a history of Haemophilia diagnosed in this center as the pregnant Haemophilia-A carrier. The management of these pregnant women was undertaken by an experienced obstetric team in this centre. The range of age in these pregnant was between 20–35-year-old. Ten ml of blood were taken, under local ethics committee approval of ICHCC, for preparation of plasma and serum samples. Some of samples were analysed immediately while others were stored at -80°C. To avoid contamination with male genetic material, all processes were performed by female personnel. Simultaneously, chorionic villus sampling (CVS) and DNA analysis have been tested in ICHCC as the gold standard method in pregnancies with a high risk of severe Haemophilia and as the positive control for this study.
DNA extraction from plasma and serum samples. Total cell-free DNA was extracted from 400 μl of plasma and serum, using the High Pure PCR Template-Preparation Kit (Roche Applied Science, Germany) according to the manufacturer’s instructions. To ensure absence of contamination during DNA extraction, a female sample was always used in each series of DNA analyses as a negative control.
Standardization of the Real-Time Duplex PCR assay. Two pairs of primers for the SRY gene, as a cell-free fetal DNA marker, and a single pair of primers for the β-actin gene used as a control for the DNA extraction process were selected [Table/Fig-1]. Positive results for amplification of beta-actin made it possible to distinguish true negative signals from the false negative results. Due to the amplification of carry-over contamination, the results can easily be vitiated, when detecting a low copy number DNA target against a high background of non-target DNA. We therefore used the dilute PCR products for real time analysis. For this purpose, the first PCR amplification was performed in a conventional thermal cycler machine, using two pairs of primers for amplifying SRY gene in a reaction volume of 25 μl containing 10x Buffer (1X), MgCl2 (2.25mM), dNTPs (0.45mM), primers (0.2μM) and Taq DNA Polymerase (1unit). After initial denaturation at 95°C for 5 minutes, the thermal cycling program was set up for 20 cycles as follows: 95°C for 45s, 56°C for 45s, 72°C for 45s. Two μl of diluted product (1: 5) was then subjected to Real-Time Duplex PCR analysis using two pairs of SRY primers in a Light Cycler 1.5 instrument, Carousel-Based System (Roche, Germany). The second Real-Time amplification reaction was performed with 1X Light Cycler® FastStart DNA Master PLUS SYBR Green and with 0.25μM of each primer in a final volume of 20 μl. Touchdown thermal cycling was started with a 10 min pre-incubation at 95°C , followed by 35 cycles of denaturation at 95°C for 10s, primary annealing at 62°C for 10s, and after 7 cycles, was then reduced (1°C/cycle ) to the secondary annealing of 56°C for 10s , extension at 72 for 10s and, at the end of PCR amplification, melting curve analysis was performed by increasing the temperature from 65°C to 95°C. During each run, a sample of male genomic DNA as the positive control, female genomic DNA as the DNA extraction negative control, and a blank sample as a PCR negative control (non-template control) were used. Results were later compared with the PCR analysis of fetal DNA samples provided by chorionic villus sampling (CVS).
Two pairs of primers for the SRY gene and one pair of primers for the B-actin are shown
| Primers | Sequences (5′→3′) | Position (Accession No.) | Size of amplicons |
|---|
| SRY-1 Forward Reverse | CAACGTCCAGGATAGAGTGA GCTGATCTCTGAGTTTCGCA | X53772.1 | 112 bp |
| SRY-2 Forward Reverse | TTGCACTGAAAGCTGTAACTC CTGTAATTTCTGTGCCTCCT | X53772.1 | 247 bp |
| B-actin Forward Reverse | AAACTGGAACGGTGAAGGTG TATAGAGAAGTGGGGTGGCT | NM_001101 | 174 bp |
Results
With amplification of any fragment of the SRY sequence, fetal sex was diagnosed as male and in the absence of such amplification, was diagnosed as female. [Table/Fig-2] shows the results of duplex/nested PCR analysis performed on male and female normal/control samples. Because of locating one pair of SRY primers within another pair, in addition to the main fragments, that is two fragments with 112 and 247 bp, two other fragments, one (169bp) resulting from coupling the second forward primer with the first reverse primer and the other (190bp), resulting from the first forward primer with the second reverse primer, will be produced. After a 20-cycle conventional PCR on the male and female controls, several diluted samples (1:10, 1:20 and 1:50 dilutions) were amplified by Real-Time PCR. Three peaks of the SRY genes with three dilutions of normal samples were observed with male genomic DNA, without any primer-dimers in the melting curve of Real-time duplex PCR, while the latter ones could be detected for female and blank samples in position with lower temperatures and without any signal for SRY as well [Table/Fig-3a] and [Table/Fig-3b]. Regarding main samples, of 23 maternal blood samples totally collected at between 8 and 12 weeks of gestation, only 15 serum could be assayed by Real-Time duplex PCR and CVS analysis [Table/Fig-4] and for the nine remaining samples, we unfortunately lost the CVS samples, it is because of some mothers wanted to keep their fetus without knowing about the fetal status and doing CVS sampling. However, comparison of Real-Time duplex PCR results with PCR analysis of fetal DNA samples provided by CVS indicated that SRY sequences were definitely detected in 6 of 6 serum samples (100% sensitivity) concerning of male samples [Table/Fig-4]. Meanwhile, SRY sequences were not detected in none of the remaining serum samples (9 cases) indicated mothers carrying female fetus (100% specificity). In female samples, only primer-dimer peaks were observed. In our experience, when we used fresh samples with male fetus, we obtained a strong SRY gene peak [Table/Fig-5a] compared with the male positive control accompany with primer-dimer peak, but when we used the same frozen/thawed samples, shorter or smaller peak of SRY ampilcon was detected in all serum samples carrying a male fetus [Table/Fig-5b]. Overall, the correct prediction rates taken as the accuracy (AC), positive predictive value (PPV) and negative predictive values (NPV) for all 15 serum samples, fresh or frozen/thawed samples, analysed in the current study were as follows: 100% (15/15), 100% (6/6) and 100% (9/9) respectively.
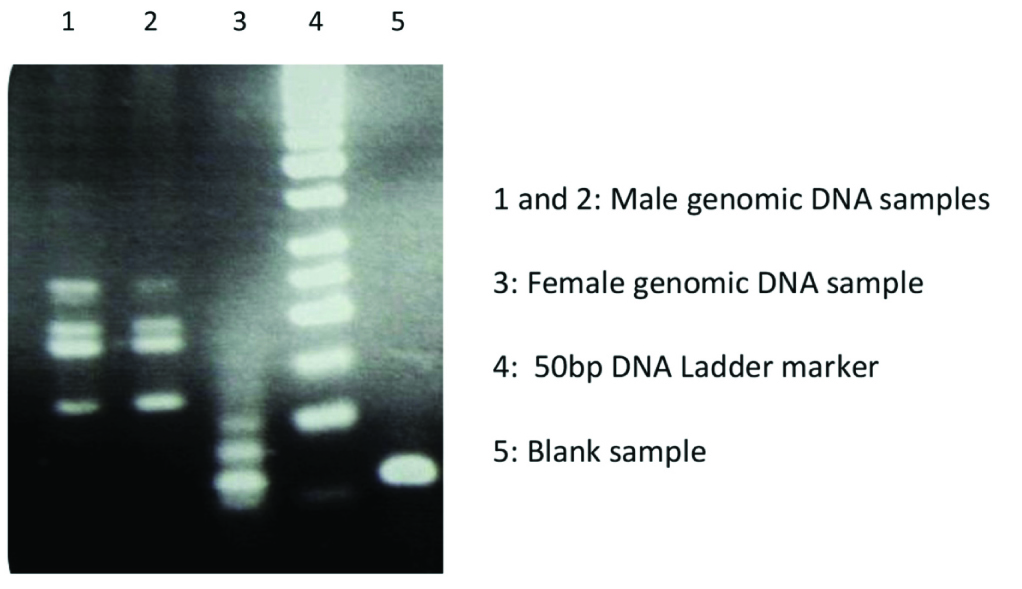
The Conventional duplex PCR on the positive and negative SRY samples
Melting curve analysis for diluted PCR product samples in the real-time duplex PCR (a) and electrophoresis result of these samples (b). Positive controls (male samples with 3 different dilutions):Peak No.1; 1:10 dilution, Peak No.2; 1:20 dilution, Peak No. 3; 1:50 dilution, Peak No. 4, for negative control or female sample; 1:50 and Peak No. 5 for Blank sample with the same dilution; 1:50.B: Agarose Gel. B: Agarose Gel electrophoresis for the results gained from the Real-Time PC

Gestational age data and results of Real-time duplex PCR on the maternal blood of Haemophilia-A carriers
| No. | Samples | The week of pregnancies | Results of Real-Time duplex PCR (SRY detected) | Results of CVS (SRY detected) |
|---|
| Plasma | Serum | Plasma | Serum |
|---|
| 1 | √ | √ | 9 | Positive | Positive | Positive |
| 2 | √ | √ | 12 | Positive | Positive | Positive |
| 3 | √ | √ | 9 | Positive | Positive | Positive |
| 4 | √ | √ | 11 | Positive | Positive | Positive |
| 5 | √ | √ | 8 | Positive | Positive | Positive |
| 6 | √ | √ | 11 | Positive | Positive | Positive |
| 7 | - | √ | 7 | - | Negative | Negative |
| 8 | - | √ | 11 | - | Negative | Negative |
| 9 | √ | √ | 9 | Negative | Negative | Negative |
| 10 | - | √ | 8 | - | Negative | Negative |
| 11 | √ | √ | 11 | Negative | Negative | Negative |
| 12 | - | √ | 11 | - | Negative | Negative |
| 13 | - | √ | 9 | - | Negative | Negative |
| 14 | √ | √ | 12 | Negative | Negative | Negative |
| 15 | - | √ | 8 | - | Negative | Negative |
Melting curve analysis of the SRY amplification of the fresh and frozen maternal samples. a) Fresh samples, and b) frozen samples

Discussion
Several noninvasive methods have been introduced for the prenatal diagnosis of fetal sex [18–21]. Due to the uncertain results of ultrasound before 13 weeks of gestation, assessment of fetal sex by this technique is usually done in the second trimester of pregnancy. Recovery of transcervical cells [22] and isolation of fetal cells from maternal blood [23] have also has been reported for the detection of fetal gender, but problems due to lack of sensitivity or specificity; frequent method failure rates and high cost make them unsuited to routine application. The detection of cell-ffDNA in maternal blood in 1997 has opened up new possibilities for noninvasive prenatal diagnosis and created an important field of study in prenatal genetic diagnosis, including fetal sex determination for couples at risk of X-linked diseases such as Haemophilia, single-gene disorders of paternal origin, and fetal RhD status in RhD- negative women. It has also been suggested that ffDNA can be used for monitoring fetal and maternal pathophysiological conditions [7,24,25]. The ffDNA can be isolated from serum and plasma of maternal blood and used for noninvasive prenatal sex determination. Ingargiola I et al., quantitatively determined ffDNA concentrations in plasma of 9 normal pregnant women carrying a male fetus, using SYBR Green in the third trimester of gestation [26]. Tungwiwat W et al., were able to determine fetal gender in 15 normal women carrying a male fetus by a multiplex nested PCR for the SRY gene [27]. Rijnders R et al., predicted fetal sex correctly in 97.8% of 45 pregnancies at risk for congenital adrenal hyperplasia using ffDNA in plasma and by Real-Time PCR and Taq Man probes, at gestational ages of 8 to17 weeks [28]. In another study conducted by this group to evaluate the earliest gestational age for detecting ffDNA in maternal plasma, SRY sequences were detected with 50% sensitivity at 5 weeks of gestation; 80% at 7 weeks and 100% at 9 weeks [29]. Hwa HL et al., predicted fetal sex in 20 of 23 healthy pregnant women carrying male fetuses. Three of samples that showed false negative results, were at a gestational age of less than 11 weeks [30]. Davalieva K et al., correctly determined the fetal gender prior to amniocentesis, in 43 of 46 maternal plasma samples (93.5%) using Real-Time PCR [31]. Zargari M et al., correctly predicted fetal sex in 40 of 42 maternal plasma samples ( 95.2%) in normal pregnant women who were in their 6th and 10th weeks of gestations using nested-PCR on the SRY gene and other genes on the chromosome Y [32]. Khorram Khorshid et al., have done the same experiments on the normal pregnant women could able to predict fetal gender with 97.3% sensitivity using real time PCR on the same genes on the chromosome Y [33]. Hromadnikova I et al., demonstrated a sensitivity and specificity of 100% in 12 pregnancies at risk of Haemophilia, 11 haemophilia A and 1 haemophilia B, and 32 pregnancies at risk of chromosomal aneuploidies in the 10th to 18th weeks of gestation using TaqMan Real-Time PCR [34]. Bustamante-Aragones A et al., assessed fetal sex with an accuracy and specificity of 100% from a gestational age of 7 weeks by Real-Time PCR analysis [35]. Two latter studies and their results were quite similar to our work, although our study differed in that we used fresh and frozen serum /plasma from the maternal blood of Haemophilia A carriers and we also used 2 pairs of primers for the SRY gene to obtain a reliable signal for fetal gender in a Real-Time duplex PCR analysis and finally, we used SYBR Green instead of a probe in this analysis. We were able to obtain a high degree of accuracy with fresh serum, rather than with plasma, which is consistence with several studies indicated that fetal DNA in serum is present in greater quantities than in plasma and its concentration increases some 100 times when samples are kept at 4°C for 4 to 5 days [36,37]. In case of the presence of the large amounts of non-target DNA which is the major problem related to amplifying low copy number DNA target sequences [38], we were well able to overcome this problem by combining duplex/nested-PCR and LC-qPCR. However, limited samples used in this experiment would be considered as lacuna of our study.
Conclusion
In this study we have tried to evaluate the specificity and sensitivity of Real-Time duplex PCR for determining fetal gender, especially for the male sex, using ffDNA in the maternal blood of Haemophilia-A carriers between the 8th and 12th weeks of gestation. The real-time duplex PCR which introduced in this study has brought up two remarkable points: the first is the specificity of this method and the second is that the best results are obtained when the fresh maternal serum taken from the Haemophilia carriers used at between 8 and 12 weeks of gestation. Meanwhile, the introduced method is not only specific, sensitive and relatively cheap, but also applicable in any well-equipped clinical laboratories for early diagnosis of fetal gender in Haemophilia-A carriers.
[1]. Peyvandi F, Jayandharan G, Chandy M, Genetic diagnosis of haemophilia and other inherited bleeding disordersHaemophilia 2006 12(3):82-89. [Google Scholar]
[2]. Mehdizadeh M, Kardoost M, Zamani G, Baghaeepour MR, Sadeghian K, Pourhoseingholi MA, Occurrence of haemophilia in IranHaemophilia 2009 15(1):348-51. [Google Scholar]
[3]. Stonebraker JS, Bolton-Maggs PH, Soucie JM, Walker I, Brooker M, A study of variations in the reported haemophilia A prevalence around the worldHaemophilia 2010 16(1):20-32. [Google Scholar]
[4]. Mujezinovic F, Alfirevic Z, Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic reviewObstet Gynecol 2007 110:687-94. [Google Scholar]
[5]. Lo YM, Recent advances in fetal nucleic acids in maternal plasmaJ Histochem Cytochem 2005 53:293-96. [Google Scholar]
[6]. Fernandez-Martinez FJ, Galindo A, Garcia-Burguillo A, Noninvasive fetal sex determination in maternal plasma: a prospective feasibility studyGenet Med 2012 14(1):101-06. [Google Scholar]
[7]. Lo YM, Chiu RW, Genomic analysis of fetal nucleic acids in maternal bloodAnnu Rev Genomics Hum Genet 2012 13:285-306. [Google Scholar]
[8]. Chan KC, Zhang J, Hui AB, Size distributions of maternal and fetal DNA in maternal plasmaClin Chem 2004 50:88-92. [Google Scholar]
[9]. Ishihara N, Matsuo H, Murakoshi H, Laoag-Fernandez JB, Samoto T, Maruo T, Increased apoptosis in the syncytiotrophoblast in human term placentas complicated by either preeclampsia or intrauterine growth retardationAm J Obstet Gynecol 2002 186(1):e158 [Google Scholar]
[10]. Bischoff FZ, Dang D, Horne C, Fetal DNA in maternal plasma circulates as apoptotic bodies: elucidation of the structural nature of fetal DNA for non-invasive prenatal genetic diagnosisAm J Hum Genet 2003 73:189 [Google Scholar]
[11]. Birch L, English CA, O’Donoghue K, Accurate and robust quantification of circulating fetal and total DNA in maternal plasma from 5 to 41 weeks of gestationClin Chem 2005 51:312-20. [Google Scholar]
[12]. Guilbert J, Benachi A, Grebille AG, Ernault P, Zorn JR, Costa JM, Kinetics of SRY gene appearance in maternal serum:detection by Real-time PCR in early pregnancy after assisted reproductive techniqueHum Reprod 2003 18:1733-36. [Google Scholar]
[13]. Honda H, Miharu N, Ohashi Y, Fetal gender determination in early pregnancy through qualitative and quantitative analysis of fetal DNA in maternal plasmaHum Genet 2002 110:75-79. [Google Scholar]
[14]. Lazar L, Ban Z, Szakacs O, Negy B, Fetal sex determination with real time PCR of fetal DNA in maternal plasmaOrv Hetil 2003 144(49):2405-09. [Google Scholar]
[15]. Zimmermann B, El-Sheikhah A, Nicolaides K, Holzgreve W, Hahn S, Optimized Real-Time Quantitative PCR Measurement of Male Fetal DNA in Maternal PlasmaClin Chem 2005 51(9):1596-604. [Google Scholar]
[16]. Jahr S, Hentze H, Englisch S, DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cellsCancer Res 2001 61:1659-65. [Google Scholar]
[17]. Lo YM, Tein MS, Lau TK, Quantitative analysis of fetal DNA in maternal plasma and serum:implications for noninvasive prenatal diagnosisAm J Hum Genet 1998b 62:768-75. [Google Scholar]
[18]. Daley R, Hill M, Chitty LS, Non-invasive prenatal diagnosis: progress and potentialArch Dis Child Fetal Neonatal Ed 2014 99(5):F426-F430. [Google Scholar]
[19]. Fan HC, Gu W, Wang J, Blumenfeld YJ, El-Sayed YY, Quake SR, Non-invasive prenatal measurement of the fetal genomeNature 2012 487(7407):320-24. [Google Scholar]
[20]. Kitzman JO, Snyder MW, Ventura M, Noninvasive whole-genome sequencing of a human fetusSci Transl Med 2012 4(137):137ra76 [Google Scholar]
[21]. Devaney SA, Palomaki GE, Scott JA, Bianchi DW, Noninvasive fetal sex determination using cell-free fetal DNA: a systematic review and meta-analysisJama 2011 306(6):627-36. [Google Scholar]
[22]. Daryani YP, Barker GH, Penna LK, Patton MA, Transcervical sampling as a means of detection of fetal cells during the first trimester of pregnancyAm J Obstet Gynecol 2000 183(3):752-54. [Google Scholar]
[23]. Bianchi DW, Fetal cells in the maternal circulation: feasibility for prenatal diagnosisBr J Haematol 1999 105(3):574-83. [Google Scholar]
[24]. Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM, Rapid clearance of fetal DNA from maternal plasmaAm J Hum Genet 1999 64(1):218-24. [Google Scholar]
[25]. Smid M, Vassallo A, Lagona F, Quantitative analysis of fetal DNA in maternal plasma in pathological conditions associated with placental abnormalitiesAnn N Y Acad Sci 2001 945:132-37. [Google Scholar]
[26]. Ingargiola I, Vaerman J, Debi`eve F, Palgen G, Free fetal DNA concentration in maternal plasma during normal labour at termPrenat Diagn 2003 23:1077-82. [Google Scholar]
[27]. Tungwiwat W, Fucharoen G, Ratanasiri T, Sanchaisuriya K, Non-invasive fetal sex determination using a conventional nested PCR analysis of fetal DNA in maternal plasmaClinica Chimica Acta 2003 334:173-77. [Google Scholar]
[28]. Rijnders R, van der Schoot CE, Bossers B, de Vroede MA, Christiaens GC, Fetal Sex Determination From Maternal Plasma in Pregnancies at Risk for Congenital Adrenal. HyperplasiaObstet Gynecol 2001 98(3):374-78. [Google Scholar]
[29]. Rijnders RJ, Van Der Luijt RB, Peters ED, Earliest gestational age for fetal sexing in cell-free maternal plasmaPrenat Diagn 2003 23:1042-104. [Google Scholar]
[30]. Hwa HL, Ko TM, Yen ML, Chiang YL, Fetal gender determination using real-time quantitative polymerase chain reaction analysis of maternal plasmaJ Formos Med Assoc 2004 103(5):364-68. [Google Scholar]
[31]. Davalieva K, Dimcev P, Efremov GD, Plaseska-Karanfilska D, Non-invasive fetal sex determination using real-time PCRJ Matern Fetal Neonatal Med 2006 19(6):337-42. [Google Scholar]
[32]. Zargari M, Sadeghi MR, Shahhosseiny MH, Fetal Sex Determination using Non-Invasive Method of Cell-free Fetal DNA in Maternal Plasma of Pregnant Women During 6(th)- 10(th) Weeks of GestationAvicenna J Med Biotechnol 2011 3(4):201-06. [Google Scholar]
[33]. Khorram Khorshid HR, Zargari M, Sadeghi MR, Edallatkhah H, Shahhosseiny MH, Kamali K, Early fetal gender determination using real-time PCR analysis of cell-free fetal DNA during 6th-10th weeks of gestationActa Med Iran 2013 51(4):209-14. [Google Scholar]
[34]. Hromadnikova I, Houbova B, Hridelova D, Replicate real-time PCR testing of DNA in maternal plasma increases the sensitivity of non-invasive fetal sex determinationPrenat Diagn 2003 23:235-38. [Google Scholar]
[35]. Bustamante-Aragones A, Rodriguez de alba M, Gonzalez-Gonzalez C, Foetal sex determination in maternal blood from the seventh week of gestation and its role in diagnosing haemophilia in the foetuses of female carriersHaemophilia 2008 14:593-98. [Google Scholar]
[36]. Lee TH, Montalvo L, Chrebtow V, Busch MP, Quantification of genomic DNA in plasma and serum samples: Higher concentration of Genomic DNA found in serum than in plasmaTransfusion 2001 41:276-82. [Google Scholar]
[37]. Umetani N, Hiramatsu S, Hoon DS, Higher Amount of free circulating DNA in Serum than in plasma is not mainly caused bye Contaminated DNA During Separation ExtreneousAnn NY Acad Sci 2006 1075:299-307. [Google Scholar]
[38]. Teo IA, Choi JW, Morlese J, Taylor G, Shaunak S, LightCycler qPCR optimisation for low copy number target DNAJ Immunol Methods 2002 270:119-33. [Google Scholar]