A few options are available for the treatment of methicillin resistant (MRSA) staphylococcal infections, such as macrolides, lincosamides and streptogramin B (MLSB) with clindamycin being one of the good alternatives, particularly for skin and soft tissue infections and work as an alternative in penicillin allergic patients [1]. However, excess and inappropriate use of MLSB agents has led to an increase in number of S. aureus strains which are resistant to MLSB as well.
In a previous study we had reported the prevalence of hospital and community associated MRSA along with antibiogram [7]. Now we undertook molecular studies for detection of erm A and erm C genes among inducible clindamycin resistant isolates, also illustrating the prevalence of MLSB resistance and antibiogram of inducible clindamycin resistance (MLSBi) and constitutive resistance (MLSBc) isolates.
Materials and Methods
The present study was done for a period of 18 months (May 2013- October 2014). During this period a total of 500 S. aureus were isolated from different clinical samples such as pus, ear swab sputum, urine, blood, swabs from different sites etc, by the standard laboratory procedures [8] in the Department of Microbiology, National Institute of Medical Sciences, Jaipur, Rajasthan, India.
Detection of MRSA: MRSA detection was done by using cefoxitin 30μg (Hi Media, Mumbai). Those isolates showed zone of inhibition less than 21mm considered as MRSA.
Disk induction test: This test was performed to detect the presence of inducible clindamycin resistance among erythromycin resistant S. aureus isolated from clinical samples. A bacterial culture suspension was made equivalent to 0.5 McFarland’s standard then a lawn culture is made on the Muller-Hinton agar plate on to which disc of clindamycin 2 μg (Hi Media, Mumbai) and erythromycin 15 μg (Hi Media, Mumbai) were placed at a distance of 15 mm edge to edge as per Clinical and Laboratory Standards Institute guidelines [9]. Four types of phenotypes were observed by the disk induction test (D-test).
Inducible MLSB phenotype: In this phenotype S. aureus isolates showed D shape zone around the clindamycin disk while resistant to erythromycin [Table/Fig-1a].
Constitutive MLSB phenotype: In this phenotype S. aureus isolates were resistant to both drugs clindamycin and erythromycin [Table/Fig-1b].
Moderate sensitive (MS) phenotype: S. aureus isolates exhibited resistance to erythromycin and sensitive to clindamycin.
Sensitive phenotype: Isolates of S. aureus sensitive to erythromycin and clindamycin.
Inducible & Constitutive MLSB
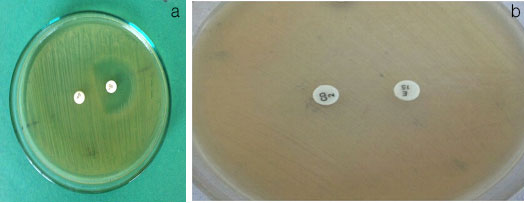
Antibiotic susceptibility test of inducible and constitutive resistant isolates
Antibiotic susceptibility test was performed by Kirby- Bauer disk diffusion method. Twelve antibiotics were used (excluding Erythromycin (15μg) and Clindamycin (2 μg)) Ciprofloxacin (5μg), Cefoxitin (30μg), Tetracycline (30 μg), Amikacin (30 μg), Gentamicin (10μg), Co- trimoxazole (25μg), Norfloxacin (10μg), Chloramphenicol (30 μg), Teicoplanin (30 μg), Nitrofurontine (300 μg), Vancomycin (30μg) and Linezolid (30μg) (Hi media Mumbai) for observation of MLSBi and MLSBc isolates [Table/Fig-2].
Antibiotic resistant pattern of MLSBi and MLSBc isolates
| Antibiotics | *MLSBi | **MLSBc | p-value |
|---|
| Ciprofloxacin | 72.22% | 67.24% | 0.67 |
| Cefoxitin | 85.18% | 55.17% | 0.011 |
| Tetracycline | 42.59% | 53.44% | 0.29 |
| Amikacin | 25.92% | 39.65% | 0.084 |
| Gentamicin | 51.85% | 37.93% | 0.14 |
| Co- trimoxazole | 37.03% | 31.03% | 0.46 |
| Norfloxacin | 25.92% | 17.24% | 0.17 |
| Chloramphenicol | 24.07% | 12.06% | 0.04 |
| Teicoplanin | 27.77% | 10.34% | 0.0035 |
| Nitrofurontine | 3.70% | 3.44% | 1 |
| Vancomycin | 0 | 0 | - |
| Linezolid | 0 | 0 | - |
**MLSBc= Constitutive resistance, *MLSBi phenotype= Inducible clindamycin resistance
erm A and erm C genes detection
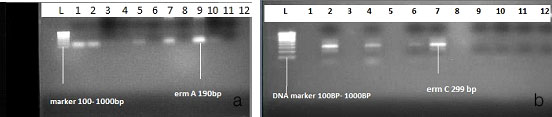
Detection of erm A and erm C genes was done by the method as described by Nizami Duran et al., [10]. Out of 54 MLSBi isolates, 24 isolates (44.44 %) were randomly selected for the observation of erm A and erm C genes which were statistically significant for evaluation of results. The genomic DNA of the selected strains was isolated by using KT- 03i (mercbioscience)
Primer designed for the study.
erm A (190 bp):
5’- AAG CGG TAA ACC CCT CTG A- ’3
5’- TTC CGC ATT CCC TTC TCA AC- ’3
erm C (299 bp) :-
5’- AAT CGT CAA TTC CTG CAT AT- ’3
5’- TAA TCC TGG AAT ACG GGT TTG- ’3
A total volume of 25 μl was taken for PCR amplification. It includes: 5 μl of genomic DNA sample which was added to 20 μl of PCR mixture. The PCR mixture consists of 20 mmol/l Tris-HCl, pH 8.4; 50 mmol/l KCl, 10 mmol/l MgCl2, and 200 μmol/l each of deoxynucleoside triphosphates (dNTPs), 0.6 μmol/l each primers and 1 U Taq DNA polymerase [10].
The amplification process was started with an initial denaturation step (95°C, 3 min). Each PCR reaction consisted of 30 cycles of amplification (denaturation at 95°C for 30 sec, annealing at 54°C for 30 sec, and DNA chain extension at 72°C for 30 sec). A final extension cycle was performed at 72°C for 4 min [10].
After amplification of the resistance genes, 10 μl of the PCR products were mixed with 3 μl of loading buffer and then loaded onto a two percent agarose gel and electrophoresis was performed in Tris-borate EDTA buffer containing 0.5 g of ethidium bromide per ml. Ethidium bromide stained DNA amplicons were visualized using a gel imaging system [10].
Results
Antibiotic resistant pattern of MLSBi and MLSBc isolates is shown in [Table/Fig-2]. Distribution of MRSA and MSSA in co-relation with various phenotypes is given in [Table/Fig-3]. Gel electrophoresis image of erm A and erm C genes among MLSBi isolates is shown in [Table/Fig-4a&b].
Distribution of MRSA and MSSA in Co-relation with various phenotypes
| Organism N= 500 | ERN-S, CLN-S (%) | ERN–R , CLN-S MS Phenotype (%) | ERN-R, CLN-R MLSBc Phenotype (%) | ERN-R, CLN-D, MLSBi Phenotype (%) |
|---|
| MRSA N= 201 | 78 (38.80) | 45 (22.38) | 32 (15.92) | 46(22.68) |
| MSSA N=299 | 221(73.91) | 44 (14.71) | 26 (8.69) | 8 (2.67) |
| TOTAL | 299 (59.8) | 89 (17.8) | 58 (11.6) | 54(10.8) |
ERN=Erythromycin, CLN= Clindamycin, S= Sensitive, R= Resistant, D= D- Shape MS=Moderate Sensitive to both drugs, MLSBc= Constitutive resistance, MLSBi phenotype= Inducible clindamycin resistance
Electrophoresis image of erm A and erm C genes among MLSBi isolates
Discussion
The D test was performed on erythromycin resistant isolates to detect the inducible phenotype. In our study we found high percentage of erythromycin resistant isolates 201(40.20%), out of them 89 (44.2%) exhibited MS phenotype, 58 (28.8%) exhibited MLSBc phenotype and 54 (26.8%) isolates were detected as inducible clindamycin resistant isolates [Table/Fig-3].
Overall prevalence of Inducible clindamycin resistance was 54 (10.8%). Kavita Prabhu et al., reported the similar results [11]. They reported 10.52% inducible clindamycin resistance among Staphylococcus aureus. Our results were also in accordance with N Seifi et al., who reported 11.37% of inducible clindamycin resistance [12]. In our study 46 (22.68%) MLSBi isolates were MRSA and only 8(2.67%) were MSSA, which is in agreement with Rahber M et al.,, who reported 22.6 % of Inducible clindamycin resistance isolates as MRSA and 4 % as MSSA [13].
We observed overall prevalence of constitute resistant isolates was 58 (11.6%) which is almost similar to the prevalence of inducible clindamycin resistant isolates. Urmi et al., reported 12% constitutive resistance among S. aureus that is similar results with our study [14]. In our study prevalence of constitutive resistant isolates among MRSA and MSSA was 32 (15.92%) and 26 (8.69%) respectively which is in accordance with K Prabhu et al., who reported 16.57 % constitutive resistance in MRSA and 6.2% in MSSA [11]. Gupta et al., reported MLSBc resistance in 19% of total isolates of which 46% were MRSA and 10% were MSSA [15].
Approximately 40 erm genes have been illustrated so far [5]. Among infection causing bacteria, erm genes are mainly borne by plasmids and transposons which are capable of being self-transferable. Therefore, a nomenclature system has been constructed to solve increasing complexity in designation. Twenty one classes of erm genes and as many corresponding erm proteins gets differentiated by this current nomenclature system. erm A, erm B, erm C, and erm F are the four major classes that are seen in pathogenic microorganisms [5,16]. The erm A and erm C determinants are predominant in staphylococci [17]. The erm A genes are mainly spread in methicillin resistant strains which are borne by transposons related to Tn554, and erm C genes are frequently responsible for erythromycin resistance in methicillin-susceptible strains that are plasmids borne, Whereas erm B class genes are mainly restricted to streptococci and enterococci, and the erm F class genes to Bacteroides species and other anaerobic bacteria [5].
Although, reports are available in which erm B has been found to be associated with MLSBi resistance among staphylococci [10,17]. Each class is relatively specific, but not strictly confined to a bacterial genus and this reveals how easily they exchange their determinants.
For the detection of erm A and erm C genes 24 randomly selected MLSBi isolates were subjected to PCR. Of the 24 isolates, 7 (29%) isolates had erm A and 3 (13%) isolates possessed erm C genes. Fourteen (58%) strains gave negative results; did not have either erm A or erm C gene [Table/Fig-4]. We observed discordance among presence of erm genes and antibiotic susceptibility, similar observation was also reported by T Zmantar et al., and Sekiguchi et al., [18,19]. Mutation in the coding or promoter region of the PCR-detected genes could be a reason for this discordance. Moreover, this result can be explained by the location of these genes in small plasmids, which were occasionally lost. There could be possibility of other erm genes like erm B, erm F, etc. among 14 negative isolates.
The prevalence of erm genes may vary area to area and the population studied. We observed much less prevalence of erm A and erm C genes when compared to other studies. In various studies prevalence of erm A was reported more than erm C like Nizami duran et al., who reported 44 (52%) erm A and 24 (28%) erm C genes [10], Lim et al., reported 82.5% erm A and 2.6% erm C gene [20]. Similarly Gul et al., also reported high prevalence of erm A genes i.e. erm A and erm C were 55 (62%) and 23 (26%) respectively [21].
However few studies reported high prevalence of erm C genes than erm A genes among MLSBi strains. 54% erm C prevalence was reported by Thakker et al., [22]. In a study conducted by Fiebelkorn et al., erm C prevalence was depicted about 71% much higher when compared to our study [23].
Conclusion
In summary, 58 (28.8%) of erythromycin resistant isolates exhibited MLSBc phenotype and 54 (26.8%) isolates were detected as inducible clindamycin resistant isolates. This reveals that about one-fourth of the erythromycin-resistant would have been bewildered as clindamycin sensitive isolates causing therapeutic failure if D-test were not performed on erythromycin resistant isolates.
We observed that resistant to erythromycin was mainly due to presence of erm A genes and there was no correlation between genotype and antibiotic susceptibility. PCR definitely has advantages over routine disk diffusion test by reducing the time for detection of resistance genes, also it is highly sensitive than routine disk diffusion test with only limitation of being little expensive. In the lights of advantages we suggest that PCR should be performed for accurate detection of genes responsible for erythromycin resistance. This kind of studies is further required in various geographical areas to fill the gap of knowledge and proper implication of antibiotics.
ERN=Erythromycin, CLN= Clindamycin, S= Sensitive, R= Resistant, D= D- Shape MS=Moderate Sensitive to both drugs, MLSBc= Constitutive resistance, MLSBi phenotype= Inducible clindamycin resistance