Haematological malignancies are immunocompromised patients who have undergone chemotherapy and received haematopoietic stem cells transplantation for treatment. Neutropenia, prolonged hospitalization, the frequent use of multiple broad-spectrum antibiotics and the use of invasive procedures all increase the risk of the nosocomial infections [1–3].
The purpose of this study was to assess the molecular characterization and resistant spectrum of Enterococci isolated from a haematology unit in a tertiary care hospital in China.
Materials and Methods
Selection of the Strains
One hundred and twenty five Enterococci strains were isolated from a haematology unit in the First Affiliated Hospital of Soochow University from September 2013 to September 2014. The haematology unit of the first affiliated hospital of soochow university has 200 beds. These strains were obtained from urine (82), blood (10), sputum (5), wound swabs (3) and others (25). The species identification of Enterococci (E. faecalis and E. faecium) was done by VITEK-2 COMPACT.
Susceptibility Testing
Antimicrobial susceptibility test for isolates of Enterococci was performed against gentamicin (GEM120, 120μg), penicillin (P, 10μg), ampicillin (AMP, 10μg), tetracycline (TCY, 30μg), ciprofloxacin (CIP, 25μg), vancomycin (VAN, 30μg), teicoplanin (TEC, 30μg) and linezolide (LZD, 30μg) (Oxoid, UK), by the disc diffusion method. MICs of vancomycin were determined by the E-test (Biomerieux, China) method on Mueller-Hinton agar. The results were interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI-2011). The vancomycin-resistant gene was identified with vanA and vanB primers. The resistant genes of aac (6′)-Ie-aph (2′′)-la, tetM and tem were identified. All the primer sequences [Table/Fig-1] have been reported in Kariyama’s studies [16].
Primers used for amplification of Resistant and Virulence genes
| Primers | Oligonucleotide sequence (5′–3′) | Sizes (bp) | Tm(°C) | Specificity | Reference |
|---|
| vanA-F | GGGAAAACGACAATTGC | 732bp | 56 | vanA | [19] |
| vanA-R | GTACAATGCGGCCGTTA | | | | |
| vanB-F | ATGGGAAGCCGATAGTC | 635bp | 56 | vanB | [19] |
| vanB-R | GATTTCGTTCCTCGACC | | | | |
| Aph-F | CCAAGAGCAATAAGGGCATA | 220bp | 56 | aac (6′)-Ie-aph(2′′)-la | [20] |
| Aph-R | CACTATCATAACCACTACCG | | | | |
| tetM-F | GTGTGACGAACTTTACCGAA | 501bp | 56 | tetM | |
| tetM-R | GCTTTGTATCTCCAAGAACAC | | | | |
| tem-F | AGGAAGAGTATGATTCAACA | 535bp | 56 | tem | |
| tem-R | CTCGTCGTTTGGTATGGG | | | | |
| cylL-L-F | AACTAAGTGTTGAGGAAATG | 159bp | 52 | cylL-L | [21] |
| cylL-L-R | AAAGACACAACTACAGTTAC | | | | |
| cylL-S-F | AGAACTTGTTGGTCCTTC | 134bp | 52 | cylL-S | [21] |
| cylL-S-R | GCTGAAAATAATGCACCTAC | | | | |
| cylL-A-F | ACAGGTTATGCATCAGATCT | 507bp | 52 | cylL-A | [21] |
| cylL-A-R | AATTCACTCTTGGAGCAATC | | | | |
| esp-F | AGATTTCATCTTTGATTCTTGG | 500bp | 50 | esp | [21] |
| esp-R | AATTGATTCTTTAGCATCTGG | | | | |
| acm-F | GGCCAGAAACGTAACCGATA | 353bp | 51 | acm | [21] |
| acm-R | CGCTGGGGAAATCTTGTAAA | | | | |
| gelE-F | AATTGCTTTACACGGAACGG | 548bp | 52 | gelE | [21] |
| gelE-R | GAGCCATGGTTTCTGGTTGT | | | | |
| asa1-F | GCACGCTATTACGAACTATGA | 375bp | 52 | asa1 | [22] |
| asa1-R | TAAGAAAGAACATCACCACGA | | | | |
| cpd-F | TGGTGGGTTATTTTTCAATTC | 782bp | 52 | cpd | [22] |
| cpd-R | TACGGCTCTGGCTTACTA | | | | |
| ace-F | GGAATGACCGAGAACGATGGC | 616bp | 52 | ace | [22] |
| ace-R | GCTTGATGTTGGCCTGCTTCCG | | | | |
DNA Isolation
All isolates were cultured on blood agar and incubated overnight at 37°. Genomic DNA was isolated from all strains with Wizard Genomic DNA purification kit (Promega), according to the manufacturer’s instructions (http://cn.promega.com/~/media/files/resources/proto cols/technical%20manuals/0/wizard%20genomic%20dna%20purification%20kit%20protocol.pdf), and used as template for PCR.
Multilocus Sequence Typing (MLST) Analysis of VRE Strains
MLST analysis was performed as described by Homan et al., [17]. The internal fragments of 7 housekeeping genes (atpA, ddl, gdh, purk, gyd, pstS and ddk) of the E. faecium isolates were amplified and sequenced. The sequence types (ST) were determined at the MLST database website (http://efaecium.mlst.net).
Detection of Virulence Genes
The genes encoding Enterococci virulence genes (cylL-L, cylL-S, cylL-A, esp, acm, gel-E, asa 1, cpd, ace), were performed by simplex PCR as reported by Sapri et al., [18]. The primers used in this study are listed in [Table/Fig-1].
Results
Bacterial Isolates and Antibiotic Susceptibility Testing
A total number of 125 Enterococci were isolated from hospitalized haematological malignancy patients. Among the isolates, 79(63.2%) were identified as E. faecalis, 46(36.8%) as E. faecium. Urine was the most common source (82, 65.6%).
The antibacterial resistant profiles of Enterococci isolates are summarized in [Table/Fig-2]. The disk diffusion indicated that the majority of Enterococci isolates were resistant to CIP (63.2%), PEN (60.8%), AMP (57.6%), TCY (51.2%) and GEH (48.0%). Among E.faecium, maximum resistance was seen against PEN 93.5% and AMP 93.5% followed by CIP 87%. They were susceptible to linezolid except one isolate of E. faecium.E. faecalis were resistant to TCY (62.0%) and CIP (49.4%). E. faecium isolates were more resistant than E. faecalis. Vancomycin resistance were detected and 8(6.4%) VREfm were found.
Molecular characterization and resistant spectrum of Enterococci
| E. faecalis (n=79, 63.2%) | E. faecium (n=46, 36.8%) | p-value | Total (n=125, 100%) |
|---|
| Resistant spectrum | | | |
| PEN | 33(41.8) | 43(93.5) | <0.001 | 76(60.8) |
| AMP | 29(36.7) | 43(93.5) | <0.001 | 72(57.6) |
| VAN | 0(0) | 8(17.4) | <0.001 | 8(6.4) |
| GEH | 37(46.8) | 23(49.8) | 0.552 | 60(48.0) |
| LNZ | 0(0) | 1(2.2) | 0.191 | 1(0.08) |
| TCY | 49(62.0) | 15(32.6) | 0.001 | 64(51.2) |
| CIP | 39(49.4) | 40(87.0) | <0.001 | 79(63.2) |
| TEC | 0(0) | 6(13.0) | 0.001 | 6(4.8) |
| Resistant genes | | | |
| tetM | 40(50.6) | 15(32.6) | 0.051 | 55(44.0) |
| Aph | 20(25.3) | 21(45.7) | 0.019 | 41(32.8) |
| vanA | 0(0) | 8(17.4) | <0.001 | 8(6.4) |
| tem | 0(0) | 0(0) | - | 0(0) |
| Virulence factors | | | |
| cylL-L | 23(29.1) | 3(6.5) | 0.002 | 26(20.8) |
| cylL-S | 29(36.7) | 2(4.3) | <0.001 | 31(24.8) |
| cylL-A | 30(38.0) | 2(4.3) | <0.001 | 32(25.6) |
| esp | 27(34.2) | 26(56.5) | 0.016 | 53(42.4) |
| acm | 25(31.6) | 42(91.3) | <0.001 | 67(53.6) |
| gel-E | 11(13.9) | 2(4.3) | 0.092 | 13(10.4) |
| asa 1 | 30(38.0) | 2(4.3) | <0.001 | 32(25.6) |
| cpd | 26(32.9) | 3(6.5) | 0.001 | 29(23.2) |
| ace | 7(8.9) | 2(4.3) | 0.310 | 9(7.2) |
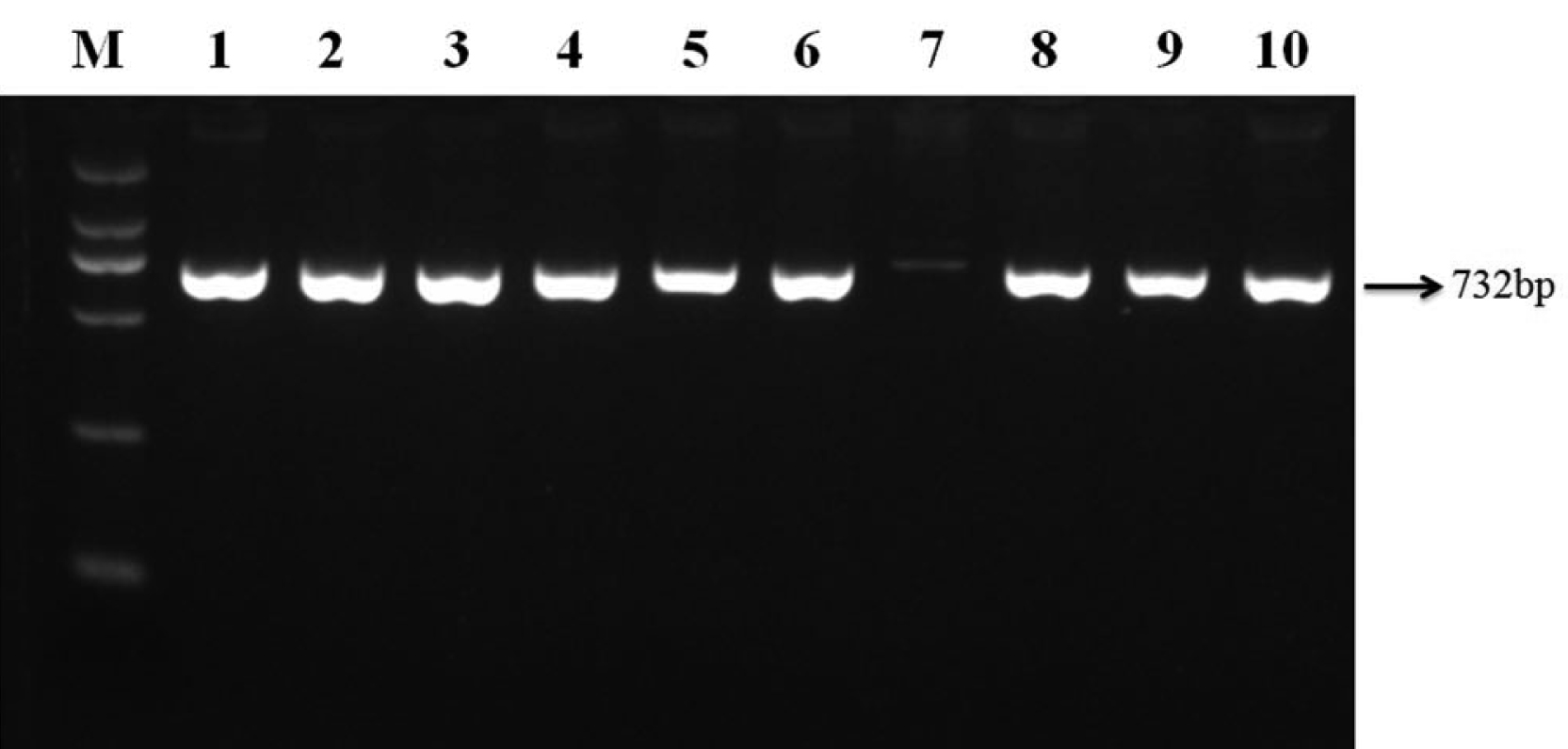
The vanA, vanB, aac(6′)-Ie-aph(2′′)-la, tetM and tem were identified. 8 strains were positive for vanA genotype and a 732-bp PCR product was obtained in all the positive isolates (data in [Table/Fig-2,3]. Forty (50.6%) E. faecalis strains were positive for tetM, and 15(32.6%) E. faecium positive for tetM (data in [Table/Fig-2]). Twenty (25.3%) E. faecalis strains were positive for aac(6′)-Ie-aph(2′′)-la, and 21(45.7%) E. faecium positive for aac(6′)-Ie-aph(2′′)-la (data in [Table/Fig-2]). However, no vanB and tem products were detected in any of the isolates.
Agarose gel electrophoresis of amplified vanA gene. Lane M: D2000 DNA Marker(100, 250, 500, 750, 1000, and 2000bp); Lanes 1-6, 8-9: Isolates positive for the vanA genes; Lane 10: positive control.
PCR Analysis of Virulence Genes
All Enterococci isolates for the presence of 9 virulence genes (VGs) was tested. The distribution of virulence gene numbers was as followed: 1 VGs (45, 36.0%), 2 VGs (32, 25.6%), 3 VGs (6, 4.8%), 4 VGs (6, 4.8%), 5 VGs (5.6%), 6 VGs (10, 8.0%), 7 VGs (3, 2.4%) and 8 VGs (3, 2.4%). 12 (9.6%) Enterococci strains did not harbor any of the tested genes. Main of these strains were isolated from non-UTI and E. faecalis (date not shown).
Of 125 Enterococci isolates, 67(53.6%) were acm, and 42.4%, 25.6%, 25.6%, 24.8%, 23.2%, 20.8%, 10.4% and 7.2% of isolates were positive for esp, cylL-A, asa 1, cylL-S, cpd, cylL-L, gel-E and ace, respectively (data in [Table/Fig-2]). The distribution of VGs among E. faecalis and E. faecium isolates entirely was shown in [Table/Fig-2].
E. faecalis isolates have more VGs than E. faecium (2.63 vs. 1.83, p<0.05). However, the most prevalent virulence determinant among E. faecium was acm (91.3%). Correlation analysis of E. faecium showed that the number of virulence factors and the resistant spectrum were negatively correlated (p<0.000). But, the number of virulence factors and the resistance spectrum of E. faecalis were never correlated [Table/Fig-3].
Characterization of VREfm
The 8 VREfm isolates showed a 100% rate of resistance to PEN, AMP and CIP. The VAN MIC values for each VREfm isolate are presented in [Table/Fig-4] and only the vanA gene was detected in all the VREfm. The esp gene was detected in 87.5% (7/8) of the isolates, and the acm gene was present in 100% (8/8) of them.
In the study, 8 VREfm isolates were subjected to MLST genotyping. Five of the 8 VREfm clinical isolates (62.5%) belonged to ST78, two to ST203 and one to ST17 (data in [Table/Fig-4]). eBURST analysis of the VREfm isolates revealed they belonged to clonal complex 17 (CC17).
Characteristics of VREfm isolates recovered from hematologic malignancy patients
| Enterococcus strain | Clinical characteristics of hematologic patients | VAN (MIC, mg/L) | Vancomycin-resistance genes | Resistant phenotype | Other resistance genes | virulence genes | ST (CC) |
|---|
| VREfm 1 | Chronic mylogenous leukaemia | >256 | vanA | PEN-CIP-VAN-TEC-AMP-LNZ | tetM | esp-acm | ST78(CC17) |
| VREfm 2 | Acute lymphocytic leukaemia | >128 | vanA | PEN-CIP-VAN-TEC-AMP | tetM | esp-acm | ST78(CC17) |
| VREfm 3 | Chronic mylogenous leukaemia | >256 | vanA | PEN-CIP-VAN-TEC-AMP-GEH-TCY | tetM-aph | esp-acm | ST78(CC17) |
| VREfm 4 | Acute lymphocytic leukaemia | >256 | vanA | PEN-CIP-VAN-TEC-AMP-GEH-TCY | tetM-aph | esp-acm | ST17(CC17) |
| VREfm 5 | Acute lymphocytic leukaemia | >128 | vanA | PEN-CIP-VAN-AMP--TCY | tetM-aph | esp-acm | ST78(CC17) |
| VREfm 6 | Mixed phenotype acute leukemia, Lung and skin infections, Broad-spectrum anti-infectious treatment | >256 | vanA | PEN-CIP-VAN-TEC-AMP-GEH | tetM-aph | acm | ST203(CC17) |
| VREfm 7 | Acute lymphocytic leukaemia | >256 | vanA | PEN-CIP-VAN-AMP-GEH-TCY | aph | esp-acm | ST203(CC17) |
| VREfm 8 | Chronic mylogenous leukaemia | >256 | vanA | PEN-CIP-VAN-TEC-AMP-GEH-TCY | tetM-aph | esp-acm | ST78(CC17) |
Discussion
Enterococci are important hospital-acquired pathogens, especially in the haematology unit. In the study, E. faecalis (63.2%) were predominant strains than E. faecium (36.8%). The species distribution is similar to that reported from different parts of the world [23,24]. The findings of multidrug resistance against the tested antibiotics were more obvious in E. faecium strains than E. faecalis strains. E. faecium strains displayed higher resistance to PEN, AMP, and CIP (≥87%). However, E. faecalis were more resistant to TCY than E. faecium (62.0% vs. 32.6%, p=0.001). The reason may be that E. faecalis is easier than E. faecium to obtain and carry genetic elements of the resistance to TCY. The result of tetM gene by PCR also confirmed this conclusion (50.6% vs. 32.6%, p=0.051).
The enterococcal surface protein (Esp) encoded by esp gene is thought to promote primary surface attachment, contributing to colonization and persistence of Enterococci in the urinary tract and biofilm formation [15]. Acm (encoded by acm), a predictor of collagen adherence, mediates E. faecium adherence to collagen [25]. Data from other paper show that Acm has contributed to the emergence of E. faecium and CC17 genotype in nosocomial infection [26]. The present study clearly revealed that E. faecalis strains carried significantly more virulence determinants than E. faecium strains (2.63 vs. 1.83, p<0.05). Esp and acm were two genes with a higher incidence in E. faecium isolates than in E. faecalis (esp: 56.5% vs. 34.2%, p=0.016; acm: 91.3% vs. 31.6%, p<0.001. The result indicates that esp and acm may make it easier for E. faecium isolates to adhere and contribute to long-term colonization.
Virulence and resistance play an important role in determining the outcome of a bacterial infection, and allow bacteria to avoid host defenses and antimicrobial treatment. The result of E. faecalis strains carrying more virulence determinants and lower resistance than E. faecium strains indicates E. faecalis isolates produce a high number of virulence factors, such as CylL, GelE, Asa1and Cpd, and introduce it in the tissues and multiply locally in spite of the host immune system. For long-term colonization and acquisition of resistance, E. faecium may reduce the introduction of the host defenses by less virulence. However, the mechanism is unclear and needs further study.
MLST is an important tool for studying the molecular epidemiology of outbreaks of E. faecium and microbial population biology [27]. In the study, MLST analysis of 8 VREfm isolates revealed three different STs: ST78, ST203 and ST17. ST17, ST78 and ST203 belonged to the clonal complex-17 (CC17) lineage, which is the cause of most of the nosocomial VRE outbreaks in Asia, Europe and Latin America, including in China [28–31]. The hospital-adapted CC17 has rapidly spread globally during the last two decades [4,28,32]. The esp gene was detected in 87.5% (7/8) of the isolates, and the acm gene was present in 100% (8/8) of them. The esp and the acm genes are associated with CC17 [33]. All VREfm patients were immunocompromised and associated with prolonged hospital stay (>60 days) and use of broad-spectrum antimicrobials. These results indicate that factors common to haematology populations (neutropenia, prolonged hospital stay and broad-spectrum antibiotic therapies) are risk factors for VRE acquisition. However, the VREfm patients had no infection symptoms, indicating that VREfm were just colonization strains. The prevalence and persistence of colonized VRE is a potential risk factor for immunocompromised patients.
Conclusion
Our data indicates that E. faecalis have different virulence factors and different resistant spectrum, compared with E. faecium, and VREfm belonged to an internationally disseminated CC17 lineage. E. faecalis isolates carry more virulence factors than E. faecium, but E. faecium isolates show more resistance than E. faecalis. The result indicates that virulence and resistance are two different mechanisms for infection or colonization of Enterococci. Molecular characterization and resistant spectrum among E. faecalis and E. faecium of haematology patients explored in this study enhanced our current knowledge of the pathogenicity and genetic characteristics of Enterococci. Moreover, determining resistant spectrum and virulence characterization is crucial for the prevention and control of the spread of nosocomial infections caused by Enterococci in the haematology unit.