Correlation Study Between HCV Genotypes Distribution Pattern and Viral Load in a Tertiary Care Hospital in Kolkata, India
Debojyoti Bhattacharjee1, Kheya Mukherjee2, Goutam Chakroborti3, Ranadeep Ghosh4, Nabarun Mandal5, Mohua Bose6
1 Assistant Professor, Department of Biochemistry, Calcutta National Medical College,32, Gorachand Road, Kolkata, West Bengal, India.
2 Assistant Professor, Department Of Microbiology,Nilratan Sarkar Medical College, Kolkata, West Bengal, India.
3 Assistant Professor, Department Of Biochemistry,Burdwan Medical College, Burdwan, West Bengal, India.
4 Assistant Professor, Department Of Microbiology, Nilratan Sircar Medical College, Kolkata, West Bengal, India.
5 Demonstrator, Department Of Biochemistry,Midnapore Medical College, Paschim Midnapore, West Bengal, India.
6 Associate Professor, Department of Microbiology, Murshidabad Medical College, West Bengal, India.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Debojyoti Bhattacharjee, 39, Russa Road, South First Lane, Kolkata, West Bengal-700033, India. E-mail : debojyoti1979@rediffmail.com
Background
Hepatitis C virus infection is a leading cause for chronic liver disease. It has wide population specific genotype variability. Genotype knowledge and viral load assessment are equally important for designing therapeutic strategies and as predictors of treatment outcome among hepatitis C (HCV) infected patients.
Materials and Methods
Between June 2012 and 2013 an observational study was conducted among 350 chronic hepatitis patients visiting Calcutta National Medical College, Kolkata, India. Among them, 110 anti-HCV antibody positive cases were diagnosed and subjected to viral RNA extraction, viral genotyping and viral load quantification using polymerase chain reaction (PCR) based techniques.
Statistical Analysis
Statistical analysis was done with IBM SPSS Statistics software, version 20. p-value <0.05 was regarded as statically significant.
Results
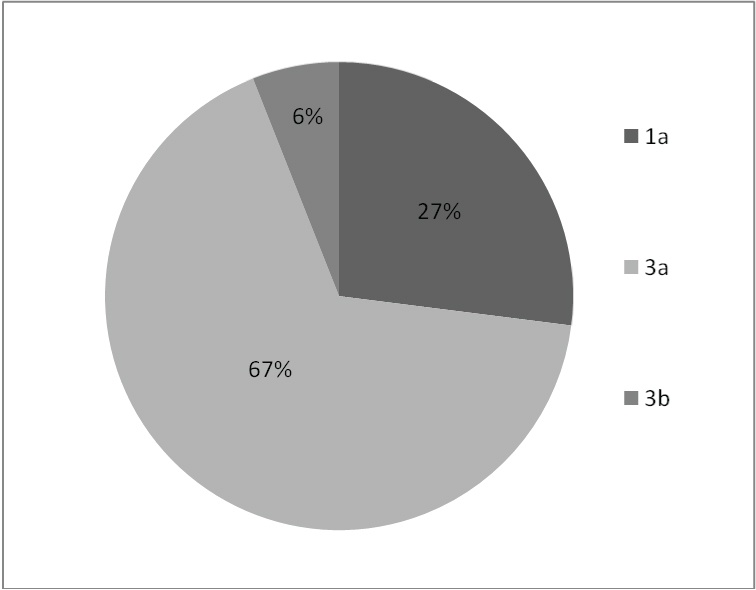
Among 66 HCV RNA positive cases, genotypes 1a, 3a and 3b were observed among 18 (27%), 44(67%) and 4(6%) cases respectively. Genotype 3a had higher viral load than patients infected with genotypes 1and 3b. However, no statistical significance was observed for viral load among the various HCV RNA genotypes.
Conclusion
Genotype 3a accounted for the highest number of cases with positive HCV RNA. However, no statistically significant difference existed for viral load among the various HCV RNA genotypes in this study.
Introduction
Hepatitis C virus infection has emerged in recent times as a leading cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma [1]. The progression of Hepatitis C virus within the liver hepatocytes more often or not is quite protracted requiring timely diagnostic interventions like serology, biochemical tests and radiological examinations for proper detection [2]. Six major HCV genotypes have been identified across the world [3]. HCV genotypes variations are important factors for patient management for these are associated with different responses to the current standard anti-HCV regimen consisting of pegaylated interferon (PEG) plus rivabarine [4]. Genotypes 1 and 4 are more resistant compared to genotypes 2 and 3 to this therapy [5]. Patient viral load also affects treatment duration and responses [6]. Patients infected with genotype 1 had higher viral loads in comparison to those with genotypes 2 or 3 [7]. However, correlation between viral loads and other HCV genotypes have not been described. The study was conducted in this background to determine the distribution pattern of HCV genotypes in plasma samples of HCV viremic individuals and their association with viral load.
Materials and Methods
The hospital-based study was conducted upon 350 patients with chronic hepatitis attending the medical outpatient department or admitted in Calcutta National Medical College, Kolkata, India in collaboration with National Institute of Cholera and Enteric Diseases (NICED), Kolkata, India during June 2012-2013. Diagnosis of cases were confirmed based upon clinical features, liver function tests, ultrasonography findings, endoscopy and wherever indicated by liver biopsy. Proper ethical committee approval was obtained from the institutional ethical committee prior to the study. Informed consent was obtained from individual patients. All the patients were diagnosed positive for HCV antibodies using 3rd generation ELISA method (J. Mitra & Co., Pvt. Ltd., New Delhi, India). Patients on immunosuppressive drugs and history of alcohol intake, evidence of HBsAg or HIV were excluded from the study.
The ELISA positive subjects were subjected to RNA extraction using QIAamp Viral RNA mini kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol.
For detection of HCV RNA, Nested RT-PCR was done based upon the 5' Un Translated Region (UTR) employing primers of Bukh et al., [8]. The positive samples gave a band at 256bp in ethidium bromide stained 1.5 % Agarose gel under a gel documentation system. HCV RNA quantification was done using ABI real time Q-RT-PCR kit (AgPath-IDTM One Step RT-PCR kit). The HCV primer and probe sequences were directed against the 5’ non-coding region of the HCV genome. HCV standards were obtained from Genome Diagnostic Pvt. Ltd., India. The HCV load in plasma was expressed as log10 international units per millilitre (log10 IU/ml). HCV genotyping was done with reverse transcriptase PCR (RT-PCR) according to Bukh and Cantaloube et al., respectively for core and NS5B in a Veriti 96 well Thermal Cycler (ABI, Foster City, USA) [9]. The snested round was performed using 2μL of the first round product.
The bands were electrophoresed using 1.5% agarose (Sigma-Aldrich, St. Louis, USA) gel and documented under gel documentation system (Bio-Rad, USA). 5’ UTR, Core and NS5B bands at 256, 405 and 389bp were excised and gel purified using QIA quick Gel extraction kit using manufacturer’s protocol [Table/Fig-1].
Nested RT-PCR amplification of 5’UTR, Core and NS5B regions of HCV genome from archived samples
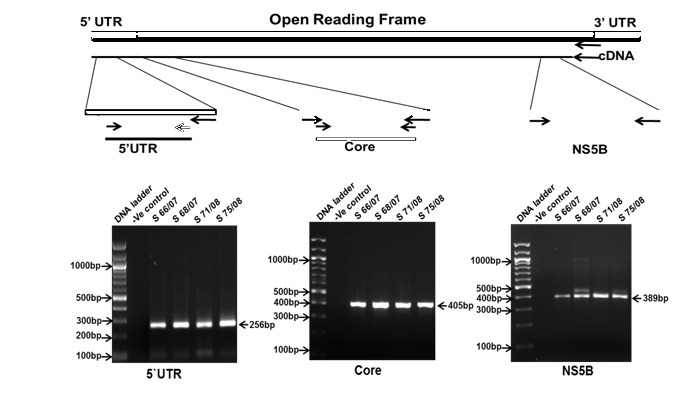
The sequencing reaction is performed using ABI big dye 3.1 sequencing kit with the manufacturer’s protocol. The reaction is performed in 10μL reaction volume with 1μL of the gel extracted product. The Sequencing PCR product is then cleaned up and loaded into the Automated DNA sequencer ABI 3130XL.
Statistical Analysis
Statistical analysis was done with IBM SPSS Statistics software, version 20. p-value <0.05 was regarded as statically significant.
Results
Of the 350 chronic liver diseases patients screened for the presence of anti-HCV antibodies, 110 were positive. These HCV antibody positive patients were tested for the presence of HCV RNA and 66 patients were found to be HCV RNA positive. All HCV RNA positive samples were subjected to genotype determination. The analysis revealed the presence of genotypes 1, 3a and 3b using RFLP and type specific PCR followed by direct sequencing. Genotype 1a was seen in 18 (27%) patients. Genotype 3 was observed in remaining 48 (73%) patients. Of these, 44 showed infection with subtype 3a (67%) while 4 had subtype 3b (6%) [Table/Fig-2].
Percentage of various HCV genotypes HCV Genotypes : 1a, 3a, 3b
Cases with mixed genotype infection were not found. Viral load quantification was carried out in all 66 HCV RNA positive patients (expressed in mean ± standard deviation) and was compared between the three groups of genotypes. The average viral load of the patients infected with genotype 3a was significantly higher than average viral load of the patients infected with genotypes 1 and 3b [Table/Fig-3]. However, no statistical significance was observed for viral load among the various HCV RNA genotypes [Table/Fig-4].
Mean viral load among HCV genotypes
| Hcv Genotypes | Number of Cases | Viral Load (Log10 Iu/Ml) (Mean ± Standard Deviation) |
|---|
| 1a | 18 | 13168.89 ± 2694.5 |
| 3a | 44 | 28733.57 ±7016.23 |
| 3b | 4 | 4774.5 ± 127.34 |
Post-hoc annova tests of multiple comparisons for viral load among HCV genotypes
| Bonferroni |
|---|
| (I) Variable 1 (HCV-genotypes) | (J) Variable 2 | Mean Difference (I-J) | Sth. Error | Significance p value * | lue |
|---|
| Lower | Upper |
|---|
| Bound | Bound |
|---|
| 3a versus | 1a | 14923.38 | 23348.16 | 1.000 | -44281.53 | 74128.30 |
| 3a versus | 3b | 23317.77 | 43579.65 | 1.000 | -87188.98 | 133824.52 |
| 3b versus | 1a | -8394.38 | 46128.00 | 1.000 | -125363.08 | 108574.30 |
* p-value significant at < 0.05
Discussion
HCV genotype distribution pattern vary widely from one continent to the other. Prevalence of genotypes 1, 2 and 3 are known to be distributed unequally throughout the world [10,11]. While Subtype 1a is prevalent in the American continents, Europe and Australia in subtype 1b prevails in North America, Europe and certain regions of Asia only [12,13]. Genotype 2 infection however is not so prevalent. [14]. HCV genotypes 1, 2 and 3 have been detected in North India, with 3 being the predominant one [15,16] while studies from South India reported high occurrence of genotype 1 followed by 3 [17,18].
The present study results showed that type 3a (66.67%) was the most common genotype followed be type 1 (27.27%) and type 3b (6.06%). No regional difference existed for HCV genotypes distribution pattern in vast regions of South Asian countries like in Iran and Pakistan where 3 was the predominant HCV genotype. However, results of given study were different from Japan and Thailand where genotype 1 was the common HCV genotype [19–21]. In our study HCV genotypes 4, 5 and 6 were not detected. These observations were similar to those reported previously regarding the near absence of these genotypes from this region [22]. These findings were similar to the conclusions obtained from other study reports from India that the most prevalent genotype is 3a followed by genotype 1a. Apart from viral genotype, viral load prior to antiviral therapy is regarded as an important prognostic sign and a valuable predictive sign for outcome of antiviral therapy. High base line viral load in terms of HCV RNA copy numbers was associated with low response to standard interferon therapy and higher probability of relapse compared to those with low-level viraemia [23]. The detected HCV genotypes and viral loads had both been extensively analysed. Many previous case reports had suggested that mean HCV RNA were higher in patients infected with genotype 1 were more likely to have higher viral loads than those infected with genotype 2 and 3. More efficient viral replication machinery of genotype 1 as compared to the others has been assumed to responsible for this [24]. However, the correlation between HCV genotypes and viral load remains controversial. In the present study the mean viral load in patients with genotype 3 was significantly higher than those with genotypes 1 and 2 but the correlation was not statistically significant (p-value>0.5). The results differ from a similar Pakistan based study where a high viral load was associated with genotypes 1a and 1b compared to other genotypes [25]. Our findings carry some important implication as timely detection and treatment are significant to achieve a high level of sustained virological response (SVR) [26]. Early time detection involves the identification of low HCV RNA level [5]. As determined by Von et al., and Dalgard et al., shorter therapy schedules for genotype 3 HCV infected patients with low baseline viral load could attain a SVR as compared to those with a high viral load [27,28]. Studies have shown that mixed HCV genotypes are more frequent in cases due to blood transfusion, especially in thalassaemic patients [26]. Franciscus had stated that mixed genotypes in a single patient may affect the antiviral therapy response and disease succession [29]. In the present study there were thalassaemic patients who had received unsafe blood in past. Fortunately, however no mixed genotype affected HCV cases were detected.
Conclusion
The present study results highlighted that genotype 3 is the predominant genotype in this geographical region followed by genotype1. However, no significant correlation was found between HCV genotypes and viral load in this study. The limitation of the study includes consideration of cases admitted only in a tertiary care hospital whereas a large number of chronic hepatitis patients are admitted in district and sub divisional hospitals. Multicentric study upon a larger population spanning over several years would have probably been helpful to further ascertain the correlation between viral genotypes and load. Our findings recommend that prior information about HCV genotype and basal RNA viral load should be an integral part of national planning strategies against HCV level at the therapeutic level. These results should help to individualize antiviral therapy, reduce side effects of antiviral therapy, economic burden and promote optimum response rates.
* p-value significant at < 0.05
[1]. Seeff LB, Hollinger FB, Alter HJ, Wright EC, Cain CM, Buskell ZJ, Long-term mortality and morbidity of transfusion associated non-A, non-B, and type C hepatitis: A National Heart, Lung, and Blood Institute collaborative studyHepatology 2001 33:455-63. [Google Scholar]
[2]. Lauer GM, Walker BD, Hepatitis C virus infectionN Engl J Med 2001 345(1):41-52. [Google Scholar]
[3]. Zein NN, Persing D, Hepatitis C genotypes: current trends and future implicationsMayo Clin Proc 1996 71:458-62. [Google Scholar]
[4]. Ahmad W, Ijaz B, Javed FT, Kausar H, Sarwar MT, Gull S, HCV genotype- specific correlation with serum markers: higher predictability for genotype 4aVirol J 2011 8:293 [Google Scholar]
[5]. Jimenez MR, Urbie SF, Guillen LP, Garza CL, Hernandez CG, Distribution of HCV genotypes and HCV RNA viral load in different geographical regions of MexicoAnnals Hepatology 2010 9:33-39. [Google Scholar]
[6]. (EASL) EAftSotL EASL Clinical Practice Guidelines: management of hepatitis C virus infectionJ Hepatol 2011 55:245-64. [Google Scholar]
[7]. Soriano V, Mocroft A, Rockstroh J, Ledergerber B, Knysz B, Spontaneous viralclearance, viral load, and genotype distribution of hepatitis C virus (HCV) in HIV-infected patients with anti-HCV antibodies in EuropeJ Infect Dis 2008 198:1337-44. [Google Scholar]
[8]. Bukh J, Purcell RH, Miller RH, Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assayProc Natl Acad Sci 1992 89(1):187-91. [Google Scholar]
[9]. Cantaloube JF, Laperche S, Gallian P, Bouchardeau F, Lamballerie X, Micco P, Analysis of the 5¢ Noncoding Region versus the NS5b Region in Genotyping Hepatitis C Virus Isolates from Blood Donors in FranceJ. Clin. Microbiol 2006 44(6):2051 [Google Scholar]
[10]. Dusheiko G, Schmilovitz-Weiss H, Brown D, McOmish F, Yap PL, Sherlock S, Hepatitis C virus genotypes: An investigation of type-specific differences in geographic origin and diseaseHepatology 2005 19:13-18. [Google Scholar]
[11]. Esfahani SHZ, Kardi MT, Edalati M, Hepatitis C virus genotype frequency in Isfahan province of Iran: a descriptive cross-sectional studyVirol J 2010 7:69-75. [Google Scholar]
[12]. Rivas-Estilla AM, Cordero-Pérez P, Trujillo-Murillo Kdel C, Ramos-Jiménez J, Chen-López C, Garza-Rodríguez Mde L, Genotyping of hepatitis C virus (HCV) in infected patients from Northeast MexicoAnn Hepatol 2008 7:144-47. [Google Scholar]
[13]. López-Labrador FX, Ampurdanés S, Forns X, Castells A, Sáiz JC, Costa J, Hepatitis C virus (HCV) genotypes in Spanish patients with HCV infection: relationship between genotype 1b, cirrhosis and hepatocellular carcinomaJ Hepatol 1997 27:959-65. [Google Scholar]
[14]. Pouillot R, Lachenal G, Pybus OG, Rousset D, Njouom R, Variable epidemic histories of hepatitis C virus genotype 2 infection in West Africa and CameroonInfect Genet Evol 2008 8:676-81. [Google Scholar]
[15]. Verma V, Chakravarti A, Kar P, Genotypic characterization of hepatitis C virus and its significance in patients with chronic liver disease from North IndiaDiagn Microbiol Infect Dis 2008 61:408-14. [Google Scholar]
[16]. Chakravarti A, Verma V, Distribution of hepatitis genotypes in β-thalassaemic patients from Northern IndiaTrans Med 2006 16:433-38. [Google Scholar]
[17]. Abraham P, Christdas J, Daniel HDJ, David J, Raghuraman S, Sivakumar J, Genotypes of hepatitis C virus in the Indian sub-continent: A decade-long experience from a tertiary care hospital in South IndiaIndian Journal of Medical Microbiolog 2013 31(4):349-53. [Google Scholar]
[18]. Valliammai T, Thyagarajan SP, Zuckerman AJ, Harrison TJ, Diversity of genotypes of hepatitis C virus in southern IndiaJ Gen Virol 1995 76:711-16. [Google Scholar]
[19]. Chowdhury A, Santra A, Chaudhuri S, Dhali GK, Chaudhuri S, Maity SG, Hepatitis C virus infection in the general population: a community-based study in West Bengal, IndiaHepatology 2003 37:802-09. [Google Scholar]
[20]. Shinji T, Kyaw Y, Gokan K, Tanaka Y, Ochi K, Analysis of HCV genotypes from blood donors shows three new HCV types 6 subgroups exist in MyanmarActa Med Okayama 2004 58(3):135-42. [Google Scholar]
[21]. Tokita H, Okamoto H, Luengrojanakul P, Vareesangthip K, Chainuvati T, Iizuka H, Hepatitis C virus variants from Thailand classifiable into five novel genotypes in the sixth (6b), seventh (7c, 7d) and ninth (9b, 9c) major genetic groupsJ Gen Virol 1995 76:2329-35. [Google Scholar]
[22]. Ahmad W, Ijaz B, Javed TF, Jahan S, Shahid I, Khan FM, Hassan S, HCV genotype distribution and possible transmission risks in Lahore, PakistanWorld J Gastroenterol 2010 16:4321-28. [Google Scholar]
[23]. Singh P, Bhatia V, Pandey M, Shashank M, Tidke P, Jha N, HCV genotypes distribution pattern & its association with viral load in indiaInternational Journal of Recent Scientific Research Research 2013 4(11):1682-84. [Google Scholar]
[24]. Ashraf A, Chakravarti A, Malik S, A study of changing trends of prevalence and genotypic distribution of hepatitis C virus among high risk groups in North IndiaIndian Journal of Medical Microbiology 2013 31(4):354-59. [Google Scholar]
[25]. Nabi SG, Zaffar G, Sheikh NI, Hassan K, Hassan U, Hepatitis C virus genotypes: A plausible association with viral loadsIndian J Pathol Microbiol 2013 56:384-7. [Google Scholar]
[26]. Pawlotsky JM, Mechanism of antiviral treatment efficacy and failure in chronic hepatitis CAntiviral Res 2003 59:1-11. [Google Scholar]
[27]. Von Wagner M, Huber M, Berg T, Peginterferon-alpha-2a (40KD) and ribavirin for 16 or 24 weeks in patients with genotype 2 or 3 chronic hepatitis CGastroenterology 2005 129:522-27. [Google Scholar]
[28]. Dalgard O, Bjoro K, Hellum KB, Treatment with pegylated interferon and ribavarin in HCV infection with genotype 2 or 3 for 14 weeks: a pilot studyHepatology 2004 40:1260-65. [Google Scholar]
[29]. Franciscus A, HCV genotypes and Quasipcies and subtypesHepatitis C support project October 2014 Version 7[http://www.hcvadvocate.org] [Google Scholar]