Several phenotypic methods are available for detection of carbapenemases like Modified Hodge test (MHT), Combined disc test (CDT) and inhibitor based E-test [Table/Fig-1]. Phenotypic methods are growth dependent, turnaround time is 18 - 24 h, not clinically useful and results are also subjective. Phenotypic tests like the modified Hodge test are useful for detection of carbapenemases but has low sensitivity [4] and low specificity [1] for NDM. Similarly for Inhibitor based Synergy phenotypic test for detection of Klebsiella pneumoniae carbapenemase, false-positive test results occur if AmpC α lactamases are coproduced [5]. Therefore, confirmation by molecular methods is necessary.
Recently, the molecular diagnostic techniques, like Real time PCR, & its modification such as LAMP have been shown to be sensitive and accurate method for identification of blaNDM-1 and blaKPC genes[6–8].
In this prospective study, we would like to evaluate various methods for detection of blaNDM-1, blaVIM, blaIMP and blaKPC genes.
Materials and Methods
This prospective study was done over a period of 9 months in department of Microbiology of Nizam's Institute of Medical Sciences. A total of 100 carbapenem resistant, clinically significant, non duplicated Gram negative isolates were included in this study (25 E. coli, 35 K. pneumoniae, 18 P. aeruginosa and 22 A. baumanii). Identification and antimicrobial susceptibility was done by Vitek 2 system, using the ID GN and the N90 AST panels.
Phenotypic tests (MHT and CDT) [9,10] were performed with all the 100 study isolates [Table/Fig-1].
CDT was done using Mueller Hinton agar (Merck) with 10 μg of meropenem (BD, USA) plain disc and with 10 μl 600μg of 3’ aminophenylboronic acid (APB) (Sigma, St.Louis, MO, USA) & 0.5 M EDTA (Himedia, India) per disk. An increase in the zone of inhibition of ≥4 mm with APB indicates presence of the KPC carbapenemase and ≥7 mm with EDTA indicates presence of an MBL.
Molecular detection of blaNDM-1, blaVIM, blaIMP and blaKPC genes
DNA extraction was done according to CDC protocol by the boiling method [11] from all the 100 isolates and the ATCC standard strains. (Commercially procured from Sterisure, Mumbai)
K. pneumoniae ATCC strain BAA1705 (positive control for blaKPC) [11] and K. pneumoniae ATCC BAA 1706 (negative control) were used.
A clinical isolate of K. pneumoniae, harboring blaNDM-1 gene, identified by PCR and gene sequencing, was included as positive control for NDM-1, because of inaccessibility of NDM-1 positive standard strain. Similarly blaVIM positive isolate was confirmed by sequencing & used as positive control.
The design of the primers for detection of blaNDM-1, blaVIM, blaIMP and blaKPC genes
For detection of blaVIM,blaIMP genes previously published primers were used, while for detection of blaNDM-1 & blaKPC genes primers were designed in house [12]. The sequences of the primers are shown in [Table/Fig-2].
Primer sequences of 3 target genes (blaNDM-1, blaVIM, blaIMP and blaKPC genes) for Multiplex PCR
| Gene | Primer Sequence (5’-3’) | Product size |
|---|
| NDM-1 FP | GCATAAGTCGCAATCCCCG | 237 |
| NDM-1 RP | CTTCCTATCTCGACATGCCG | |
| VIM FP | GTTTGGTCGCATATCGCAAC | 382 |
| VIMRP | AATGCGCAGCACCAGGATAG | |
| IMP FP | GAAGGCGTTTATGTTCATAC | 587 |
| IMP RP | GTAAGTTTCAAGAGTGATGC | |
| KPC FP | TCGAACAGGACTTTGGCG | 201 |
| KPC RP | GGAACCAGCGCATTTTTGC | |
Procedure of the Multiplex PCR assay: A 237 bp region of blaNDM -1, 382 bp region of blaVIM, 587 bp region of blaIMP & 201 bp region of blaKPC gene were amplified through the Multiplex PCR using NDM-1, VIM, IMP & KPC specific primers (synthesized at Active oligos, ILS, Gurgaon, India).
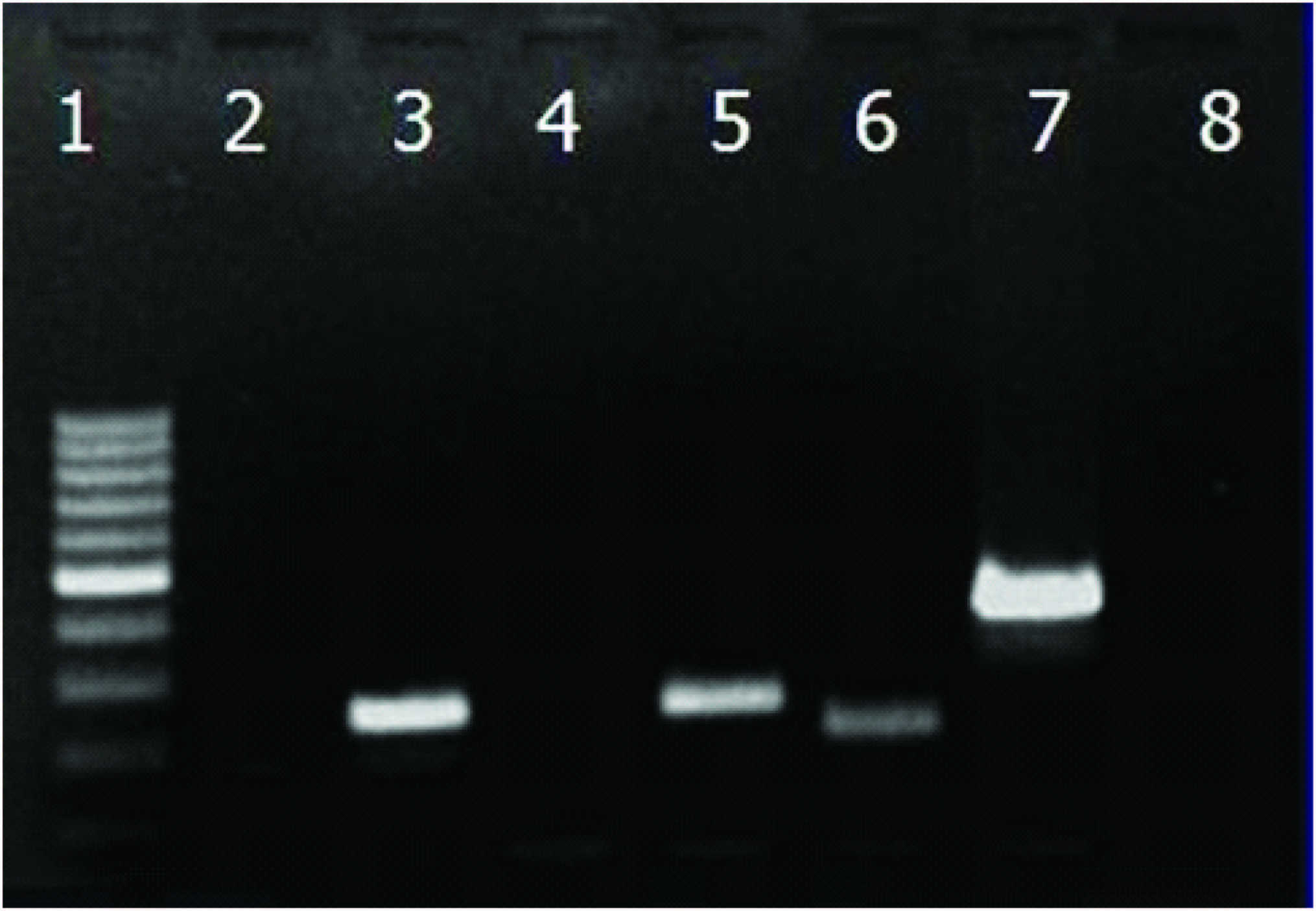
The Quick-load Taq 2X PCR Master Mix (New England BioLabs, Inc) was used, 1x PCR contains 10mM Tris-HCL(pH 8.6, @25° C), 50 mM KCL,1.5 mMMgCl2, 0.2 mM of each dNTP, 5% glycerol, 0.08% NP-40, 0.05% Tween-20, 0.024% Orange G, 0.0025% Xyelene Cyanol FF, 50 units/ml Taq DNA polymerase and nuclease-free water to make up the final volume (25 μl). Thermal cycling (Perkin Elmer, USA) for 30 cycles was done at 94°C for 1 min, 54°C for 1 min and 72°C for one and half min. And the final extension step was performed for 5 min at 72°C. The PCR product containing amplicons was analysed in a 2% agarose gel in 1x TAE buffer at 80 V for 1.5 hr and was visualized with ethidium bromide using a gel documentation system (Syngene, UK) [Table/Fig-3].
Agarose gel results of KPC, NDM-1, VIM and IMP
(Well 1: 100bp ladder (fermentas), Well 2: Negative control, well 3: NDM-1 positive sample, Well 4: Negative sample, Well 5: NDM-1 positive control, Well 6: KPC positive control, Well 7: VIM positive control, Well 8: Negative sample)
Results
Results of Phenotypic methods
Out of the 100 carbapenem resistant isolates, 70 isolates were MHT positive, while 65 isolates were CDT positive. Five isolates which were MHT positive but CDT was negative, none of the 4 genes were detected. Correlation of MIC and carbapenemase production among E. coli, K. pneumoniae, P. aeruginosa and A. baumannii is shown in [Table/Fig-4].
Correlation of MIC with carbapenemase production among GNB
| Organism & carbapenemase | Imipenem MIC (μgm/ml) | Meropenem MIC (μgm/ml) |
|---|
| 2 | 4 | 8 | 16 | 64 | 2 | 4 | 8 | 16 | 64 |
|---|
| VIM (6) |
| E. coli (2) | - | - | 1 | 1 | - | | | - | 1 | 1 |
| A. baumannii (1) | - | - | - | 1 | - | | | - | - | 1 |
| P. aeruginosa (3) | 1 | 2 | - | - | - | | 1 | 1 | 1 | - |
| KPC (15) |
| E. coli (1) | 1 | - | - | - | - | - | - | 1 | - | - |
| K. pneumoniae (5) | - | - | 1 | 4 | - | - | - | - | 2 | 3 |
| A. baumannii (9) | - | - | - | 8 | 1 | - | - | - | - | 9 |
| NDM-1 (59) |
| E. coli (14) | 2 | 4 | 6 | 2 | - | - | 3 | 4 | 5 | 2 |
| K. pneumoniae (30) | - | 3 | 8 | 19 | - | - | - | 3 | 8 | 19 |
| A. baumannii (15) | - | - | - | 14 | 1 | - | - | - | - | 15 |
Results of Genotypic methods
The results of the Multiplex PCR for four target genes are shown in [Table/Fig-5]. Out of 100 carbapenem resistant isolates, 65 isolates harboring one or more than one genes, while in 35 isolates none of the gene was detected. The most common resistance gene was blaNDM-1 (59/100) followed by blaKPC (15/100) while the blaVIM gene was least frequent (6/100). We didn’t find blaIMP in any of the isolates. Correlation of Multiplex PCR with MHT and CDT among carbapenemase producing isolates is mentioned in [Table/Fig-6].
Results of Genotypic test (Multiplex PCR)
| Organism | VIM | IMP | NDM-1 | KPC | VIM & NDM-1 | KPC & NDM-1 | Total |
|---|
| E. coli | 2 | - | 12 | 1 | - | 2 | 17 |
| K. pneumoniae | - | - | 25 | - | - | 5 | 30 |
| A. baumannii | - | - | 7 | - | 1 | 7 | 15 |
| P. aeruginosa | 3 | - | - | - | - | - | 3 |
| Total | 5 | - | 44 | 1 | 1 | 14 | 65 |
Correlation of Multiplex PCR with MHT and CDT among carbapenemase producing isolates
| Organism | KPC positive (15) | NDM -1 positive (59) | VIM positive (6) |
|---|
| PCR positive | MHT positive | CDT positive | PCR positive | MHT positive | CDT positive | PCR Positive | MHT positive | CDT Positive |
|---|
| E.coli (25) | 3 | 3 | 3 | 14 | 12 | 14 | 2 | 2 | 2 |
| K. pneumoniae (35) | 5 | 5 | 5 | 30 | 30 | 30 | - | - | - |
| A. baumannii (22) | 7 | 7 | 7 | 15 | 15 | 15 | 1 | 1 | 1 |
| P. aeruginosa (18) | - | - | - | - | - | - | 3 | 3 | 3 |
Discussion
Resistance of Carbapenem agents is due to carbapenemase and presence of other resistance mechanisms, such as ESBLs, porin mutations and/or presence of efflux pumps [13]. In our study 65 isolates were carbapenemase producers while 35 isolates were negative suggesting resistance mechanism other than carbapenemase production.
Accurate susceptibility data is required to provide effective therapy. However, automated susceptibility systems may be unreliable for detection of carbapenem resistance [14,15]. A review of several automated systems showed that they incorrectly labeled up to 87% of carbapenemase-producing K. pneumoniae isolates as susceptible to imipenem, as well as reporting varying susceptibilities for the same isolate from day to day [14]. Ertapenem resistance seems to be a marker for carbapenemase production when automated testing methods are used [14,15]. This necessitates the need for further testing by Phenotypic & genotypic methods. If resources are limited, an elevated MIC for ertapenem could be used as a screening method to determine which isolates need further testing [14,15].
Carbapenem MICs for Carbapenemase producing isolates may vary within a broad range of values, from 0.12 to >256 mg/L [16,17]. Although VIM enzymes have strong carbapenem- hydrolytic activity, a proportion of VIM-producing K. pneumoniae isolates have low carbapenem MICs. In Our sudy 50% of VIM producing isolates had an MIC ≤ 4 mg/L [17]. In contrast, isolates producing the NDM – 1 have higher carbapenem MICs, 71% of our isolates have MIC ≥ 16 mg/L [18]. Of the total carbapenemase producing isolates, most resistant isolates were A. baumannii, all of which had MIC ≥ 16 mg/L.
Phenotypic methods like MHT give variable results. MHT performed well for KPCs and OXA-48-like enzymes but poorly for NDMs, VIMs, and IMPs [19]. Only 66% of MBL producing isolates of P. aeruginosa and Acinetobacter spp. gave positive results by the MHT (Lee et al.,) in the same study 10 more isolates with equivocal results became positive with incorporation of zinc sulfate [20].
In 5 MHT positive and CDT negative isolates none of the genes included in our study were amplified, which can be explained by presence of blaOXA genes. There is currently no phenotypic test capable of detecting OXA-48. This again necessitates the need of Molecular assay.
Compared to MHT, CDT is a satisfactory and inexpensive method for detection and characterization of the carbapenemase, as results are very well correlated with PCR. Considering PCR as the gold standard test, our data suggest, CDT has 100% sensitivity and specificity.
There are very few available data of KPC from India [21]. To the best of our knowledge, our study is the first report on Multiplex PCR for detection of blaNDM-1, blaVIM, blaIMP and blaKPC genes among E. coli, K. pneumoniae, P. aeruginosa & A. baumannii and second report for the combined detection of blaNDM-1 and blaKPC genes from India [21].
We found blaKPC genes among 14 isolates with blaNDM-1 and in one isolate as a lone gene. Overall, sensitivity and specificity of MHT is 58% and 93%. However The PCR had 100% sensitivity and specificity [19]. We recommend molecular methods like Multiplex PCR for the optimal detection of carbapenemase.
Conclusion
Our results suggest that the CDT should be preferred over the MHT for the detection of carbapenemases. The Multiplex PCR was found to be more sensitive than existing phenotypic methods. Multiplex PCR will also help in simultaneous detection of various genes, reducing material, manpower & cost. It helps in determining epidemiology related to these genes & infection control.