Materials and Methods
Study design: Prospective study at a private microbiology laboratory in Mumbai, India.
Study group & period: Yeast isolates, recovered from clinical specimens, during a period of 25 months, were included in the study. Clinical relevance was not separately ascertained after isolation, as all specimens had been received for fungal culture, from patients with suspected fungal disease. Main aim of our study was to evaluate identification methodologies, and not establish clinical relevance. Isolates from repeat specimens of same patients were not included.
Materials
Sabaraud Dextrose agar (SDA) and Brain Heart Infusion agar (BHI): prepared in-house using commercial dehydrated media.
CHROMagarTMCandida (CC) (M/S BD BBLTM ,Becton Dickinson & co. Sparks, MD, USA).
API ID 32C Identification test (API) (Biomerieux, Marcy-l’Etoile, France).
Methods
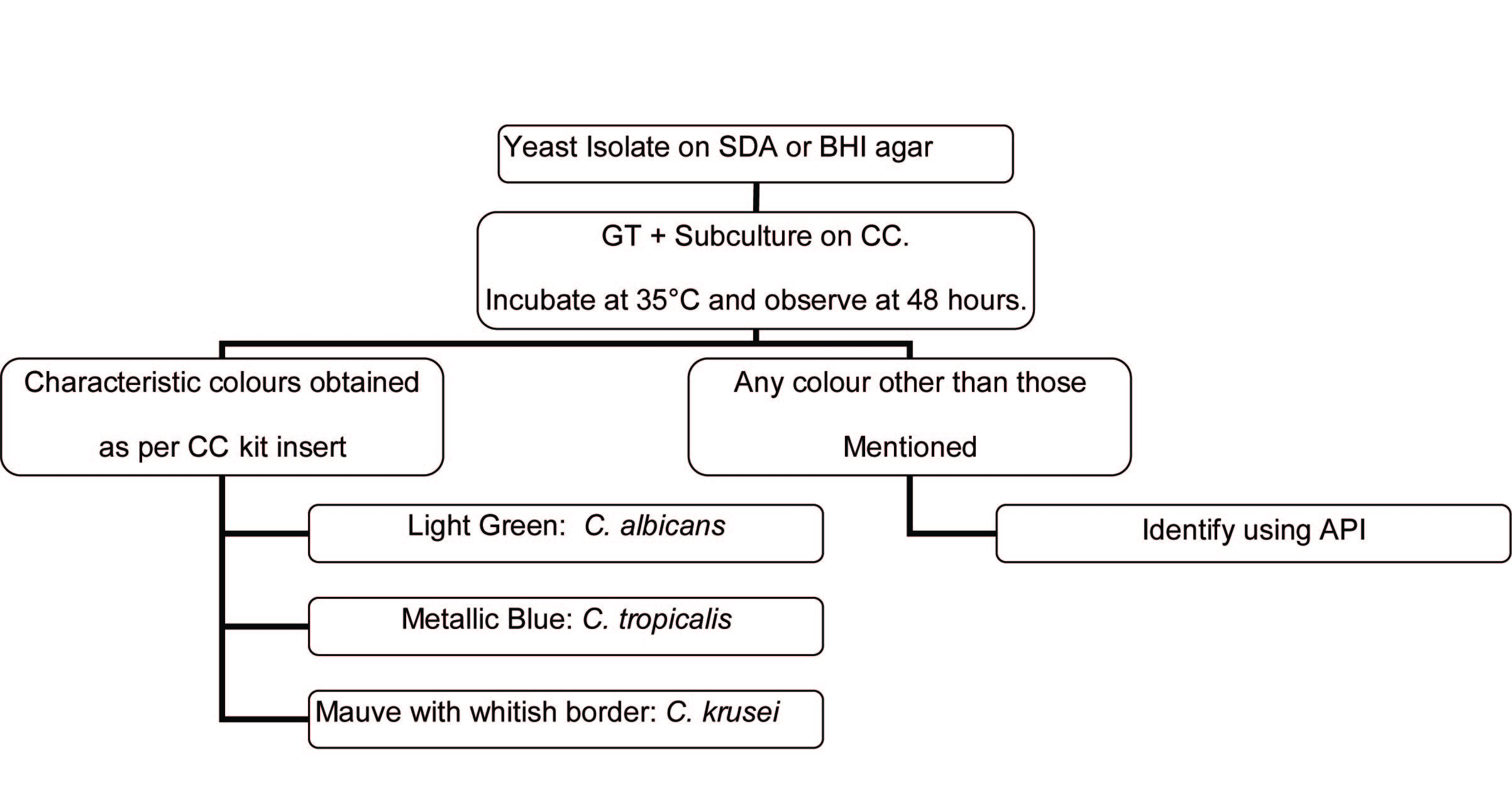
The identification algorithms shown in [Table/Fig-1&2] were used to identify the isolates.
Identification algorithm for blood culture isolates
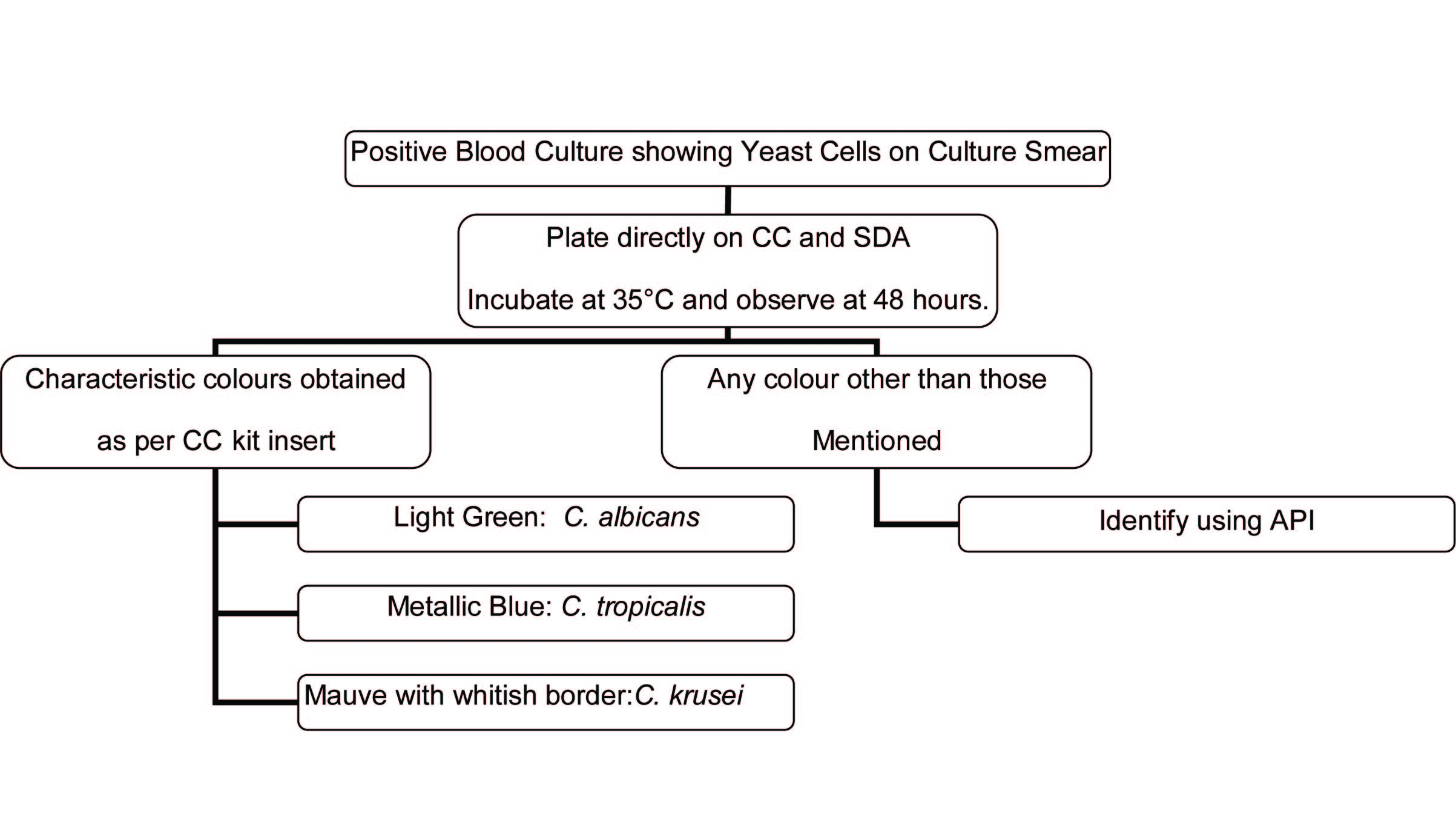
Identification algorithm for isolates recovered on solid media
Quality Control(QC):
SDA and BHI: New lot QC for its sterility and ability to support growth.
GT: Daily QC with positive and negative controls (ATCC strains) to check for sera reactivity.
CC: Prior to initiating the study, manufacturer defined colour reactions were verified for accuracy and reproducibility, using ATCC strains. Subsequently, new lot QC was done on a routine basis, as mentioned in [Table/Fig-3].
API: New lot QC
ChromagarTMcandida quality control
| Strain | Expected Colour Reaction |
|---|
| Candida albicans ATCC. 10231 | Light green |
| Candida tropicalis ATCC 1369 | Metallic blue |
| Candida krusei ATCC 6258 | Light mauve to mauve, flat with whitish border |
Results
Of 6375 clinical specimens screened during the study period, 526 clinical specimens yielded yeasts on culture. Positive specimen types included Sputum(161) ,BAL(92),Urine(83),Blood(76),Tracheal secretion(31),Endotracheal secretions(17), CSF(16), Tissue(15), Pus(10), Fluids(7),Stool(7),Body fluids(6), Ear swabs(3) & Bone marrow(2) .
Seven (4 sputa and 3 urine) of the 526 specimens showed mixed cultures, with growth of 2 types of yeasts. Hence, a total of 533 isolates were obtained. The identification profile of the isolates has been shown in [Table/Fig-4].
Identification profile (N= 533)
| Isolates | Number |
|---|
| C. albicans | 285 |
| C. tropicalis | 117 |
| C. parapsilosis | 24 |
| C. glabrata | 20 |
| C. krusei | 19 |
| Trichosporon spp. | 18 |
| C. sake | 14 |
| C.neoformans | 10 |
| C. lipolytica | 5 |
| Rhodotorula spp. | 4 |
| C. rugosa | 3 |
| C. kefyr(pseudotropicalis) | 3 |
| C. pelliculosa | 3 |
| C. dublinensis | 3 |
| Geotricum spp. | 2 |
| C. intermedia | 1 |
| C.utilis | 1 |
| C. inconspicua | 1 |
| Total | 533 |
Discussion
Identification profile of Candida & other yeast isolates
C. albicans, C. tropicalis, C parapsilosis and C. glabrata were the most commonly isolated yeasts in the study. Interestingly, a significant proportion of unusual isolates were recovered, whose identification can alter patient management. It is known that mere isolation of yeasts from clinical specimens does not establish their role in disease. Patient histories as well as repeated isolation from same site are needed to establish clinical relevance.
The diverse identification profile obtained in the present study substantiates the need for all diagnostic microbiology laboratories to be better prepared for identifying unusual yeasts. Correct identification itself may also aid in determining the significance. Larger clinical studies are also recommended to establish the clinical relevance of these unusual yeasts in the Indian context.
Utility of CHROMagarTMCandida (CC)
The use of CC allowed identification of 7 specimens containing mixed yeast species, an advantage shared by other investigators also [7,8]. None of these mixed cultures were evident on the primary culture media and were detected, only after subculturing on CC. In fact, some researchers have found CC to be as good as SDA for primary isolation of yeasts and superior to SDA in terms of suppressing the bacterial growth and time to positivity [7,8].
All 285 isolates of C. albicans gave a distinctive light green colour on CC [Table/Fig-5] The colour production was unique and was not shared by any other species. A number of studies have already substantiated this finding, reporting sensitivity & specificity of 98-100 & 100 % respectively [6,9–14]. C. dublinensis was found to give characteristic dark green colonies, distinctive from those of C. albicans. This finding has also been shared in a study by Mary-Ann et al., [15] and has been attributed to the reformulation of the CC by the manufacturer. Another study suggests that the dark green colour is found to be more pronounced, if plates are incubated beyond 48 h [10]. Majority of C. tropicalis isolates, i.e. 90.7 % (107/118) showed typical metallic blue colonies.10 isolates were identified using API, as the colours were not comparable to that produced by the C. tropicalis ATCC strain. Similar to present study, others have reported sensitivity and specificity rates for C. tropicalis, between 66.7-99% and 93.8-100% respectively [11,14].
Association of colour on CC with isolate identification
| Isolate (n=533) | Colour on CHROMagarTMCandida |
|---|
| Green | Blue | Mauve | Cream | Yellow orange |
|---|
| Light | Dark | Metallic Blue | Light Blue Rough | Glossy | Rough with white border | Mauve | Pink Brown glossy | Glossy | Rough |
|---|
| C. albicans | 285 | - | - | - | - | - | - | - | - | - | - |
| C. tropicalis | - | - | 107 | 10 | - | - | - | - | - | - | - |
| C. parapsilosis | - | - | - | - | - | - | - | - | 24 | - | - |
| C. glabrata | - | - | - | - | 14 | - | 4 | 2 | - | - | - |
| C. krusei | - | - | - | - | - | 2 | 16 | - | - | 1 | - |
| T. asahii | - | - | - | 12 | - | - | - | - | - | 4 | - |
| T. mucoides | - | - | - | - | - | 1 | - | 1 | - | - | - |
| C. sake | - | - | - | - | - | - | - | 14 | - | - | - |
| C.neoformans | - | - | - | - | - | - | - | - | 10 | - | - |
| C. lipolytica | - | - | - | - | - | - | - | - | 5 | - | - |
| Rhodotorula spp. | - | - | - | - | - | - | - | - | - | - | 4 |
| C. rugosa | - | - | - | 3 | - | - | - | - | - | - | - |
| C. pelliculosa | - | - | - | - | 2 | - | 1 | - | - | - | - |
| C. dublinensis | - | 3 | - | - | - | - | - | - | - | - | - |
| C. kefyr | - | - | - | - | - | - | - | 1 | 1 | 1 | - |
| G. candidum | - | - | - | - | - | - | - | - | 2 | - | - |
| C. inconspicua | - | - | - | - | - | - | - | - | 1 | - | - |
| C. utilis | - | - | - | - | 1 | - | - | - | - | - | - |
| C. intermedia | - | - | - | - | - | - | - | - | 1 | - | - |
The high reliability of CC to identify C. albicans and C. tropicalis provided rapid identification, thereby enabling faster reporting and timely patient management. CC was especially helpful in identifying blood isolates, as blood from yeast-positive Blood Culture bottles was directly plated on CC. This finding has been shared by Ainscough et al., [12].
Though CC is standardized for identification of C. krusei, only 2 out of 19 isolates gave characteristic mauve coloured colonies with whitish border, those matching the ATCC C. krusei strain. 16 isolates gave mauve coloured colonies without whitish border and were subsequently identified using API. Review of literature, also shows conflicting reports about the sensitivity of C. krusei detection on CC. Some studies have claimed a 100% sensitivity and specificity in their study [6,9,13]. However, there could be a bias towards CC in some studies, as the authors have used API confirmed isolates for evaluating CC, and not looked at CC as the first line of identification. Also, in studies where CC has been used as primary identification medium, identities of the “C. krusei” flat mauve colonies have not been subsequently confirmed using confirmatory identification [5].
Certain reports suggest CC’s utility in reliably identifying C. glabrata, while others have discussed issues with colour intensity of C. glabrata strains [10,11]. Findings in present study corroborate with the latter group, as characteristic mauve glossy colonies were observed only in 14 of the 20 C. glabrata isolates. Also, 2 strains of C. pelliculosa and 1 strain of C. utilis produced similar colonies. Odds et al., [14] have also reported pink to purple coloured colonies of C. pelliculosa on CC.
12 out of 16 strains of T. asahii and 3 C. rugosa isolates demonstrated light blue coloured rough colonies on CC. , a finding also described by Paritpokee et al., [9]. All 24 isolates of C. parapsilosis in the present study showed cream coloured colonies, a finding shared by Vijaya D et al., [8]. However, cream coloured colonies were noted in a number of species and were not specific to C. parapsilosis.
Thus, certain characteristic colony morphologies on CC were found to be associated with C. glabrata, T. asahii and C. sake .Though present study data indicates that a presumptive identification of these species can be deduced from the typical morphology, these characteristics are not enough to conclude on the isolate identity. It is worthwhile to mention, that morphology on corn meal agar, if performed in addition to plating on CC, might help in reliable identification of certain species, especially C. glabrata and T. asahii.
Usefulness on the Yeast Identification Algorithm
Reliable identification of 393/533, i.e. 73.7% of isolates was possible using GT and CC Tests only [Table/Fig-6]. This was because C. albicans and C. tropicalis, which can be very reliably, identified using chromogenic media, constituted 75.2 % of the clinical isolates in our study.
Usefulness on the yeast identification algorithm
| Light Dark | No. | Identification established by |
|---|
| % | GT+CC | GT+CC+API | % |
|---|
| Germ tube positive isolates(n=287) | C. albicans | 284 | 284 | 100 | 0 | 0 |
| C dublinensis | 3 | 0 | 0 | 3 | 100 |
| Germ tube negative isolates(n=246 | C. albicans | 1 | 0 | 0 | 1 | 100 |
| C. tropicalis | 117 | 107 | 91.45 | 10 | 8.55 |
| C. parapsilosis | 24 | 0 | 0 | 24 | 100 |
| C. glabrata | 20 | 0 | 0 | 20 | 100 |
| C. krusei | 19 | 2 | 10.53 | 17 | 89.47 |
| Trichosporon spp. | 18 | 0 | 0 | 18 | 100 |
| C. sake | 14 | 0 | 0 | 14 | 100 |
| C. neoformans | 10 | 0 | 0 | 10 | 100 |
| C. lipolytica | 5 | 0 | 0 | 5 | 100 |
| Rhodotorula spp. | 4 | 0 | 0 | 4 | 100 |
| C. pelliculosa | 3 | 0 | 0 | 3 | 100 |
| C. kefyr | 3 | 0 | 0 | 3 | 100 |
| C. rugosa | 3 | 0 | 0 | 3 | 100 |
| Geotricum spp | 2 | 0 | 0 | 2 | 100 |
| C. intermedia | 1 | 0 | 0 | 1 | 100 |
| C. inconspicua | 1 | 0 | 0 | 1 | 100 |
| C. utilis | 1 | 0 | 0 | 1 | 100 |
| Total | 533 | 393 | 73.73 | 140 | 26.27 |
Identity of 140 isolates, i.e. 26.3 %, could not be confirmed on the basis of GT and CC tests only. Identification of these isolates was performed using API ID 32C. It is thus evident that GT or CC testing cannot suffice, for accurate identification of all yeast species, and must be accompanied by more thorough identification protocols.
The cost of a single CC plate is approximately 1/10th that of a single API 32 C strip. Hence, if CC and API are used in a step-wise identification algorithm, yeast identification can be performed in a more cost effective manner.
Conclusion
The diverse identification profile obtained in the present study substantiates the need for all diagnostic microbiology laboratories to be better prepared for identifying unusual yeasts and thus facilitate correct treatment to minimize treatment failure or recurrent infections .Larger clinical studies are recommended to establish the clinical relevance of unusual yeasts in the Indian context.
CC is a valuable and cost effective tool for 1) identifying C. albicans and C. tropicalis isolates, 2) detecting mixed cultures and 3) rapid identification of blood culture isolates. GT or CC testing cannot suffice for identification of all clinically encountered yeasts. However, use of GT, CC and automated identification systems in a step-wise algorithm can enable the same in a more cost effective manner.