Many reports are available in the literature regarding the identification of vancomycin-resistant Enterococci (VRE) using conventional microbiological methods, which require time, resource and space. These standard methods are labour-intensive and require 48-72 h to give the result. Therefore, management of VRE infection relies on rapid and sensitive detection [6].
Chromogenic media are increasingly used as versatile tools in early differentiation and identification of VRE from clinical samples [7]. The present study was undertaken to evaluate the two different chromogenic media, CHROMagarTM VRE (France) and Hicrome VRE (Himedia, India) in detecting VRE in comparison with E- test (Himedia, India).
Materials and Methods
The present study was carried out in the Department of Microbiology, Adichunchanagiri Institute of Medical Sciences, BG Nagara from June 2013 to May 2014. The ethical committee of the institution granted approval for the study. Out of 4489 clinical samples screened, 100 were Enterococci. Isolates were identified and speciated. Further confirmation was done using Group D anti-sera (Histrep, Himedia India) and CHROMagarTM orientation agar (CHROMagar France).
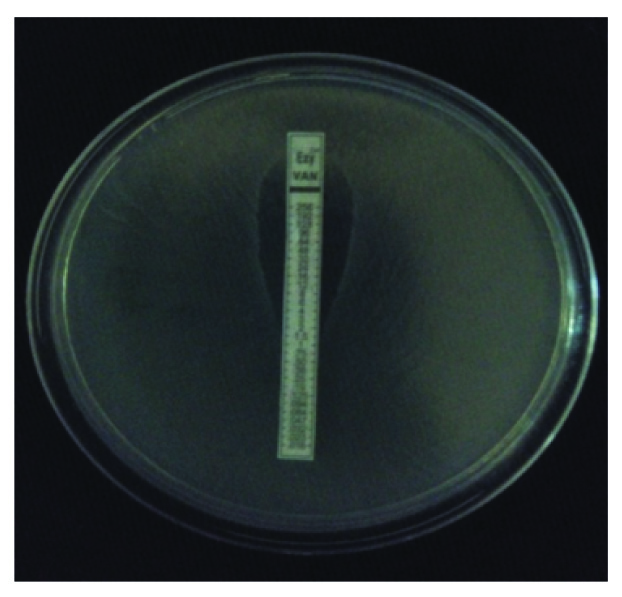
The minimum inhibitory concentration (MIC) for vancomycin is determined by E-test as shown in [Table/Fig-1] (0.016-256 μg/ml). The MIC ≥ 32μg/ml is considered as VRE based on the CLSI guidelines [8].
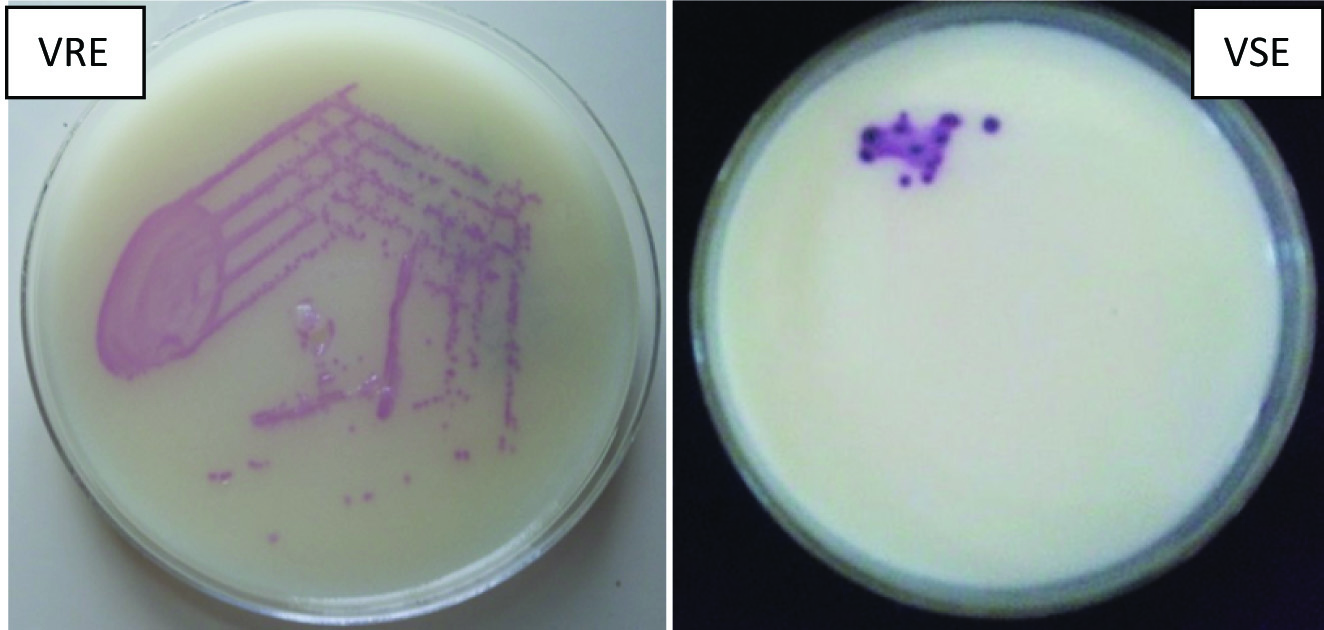
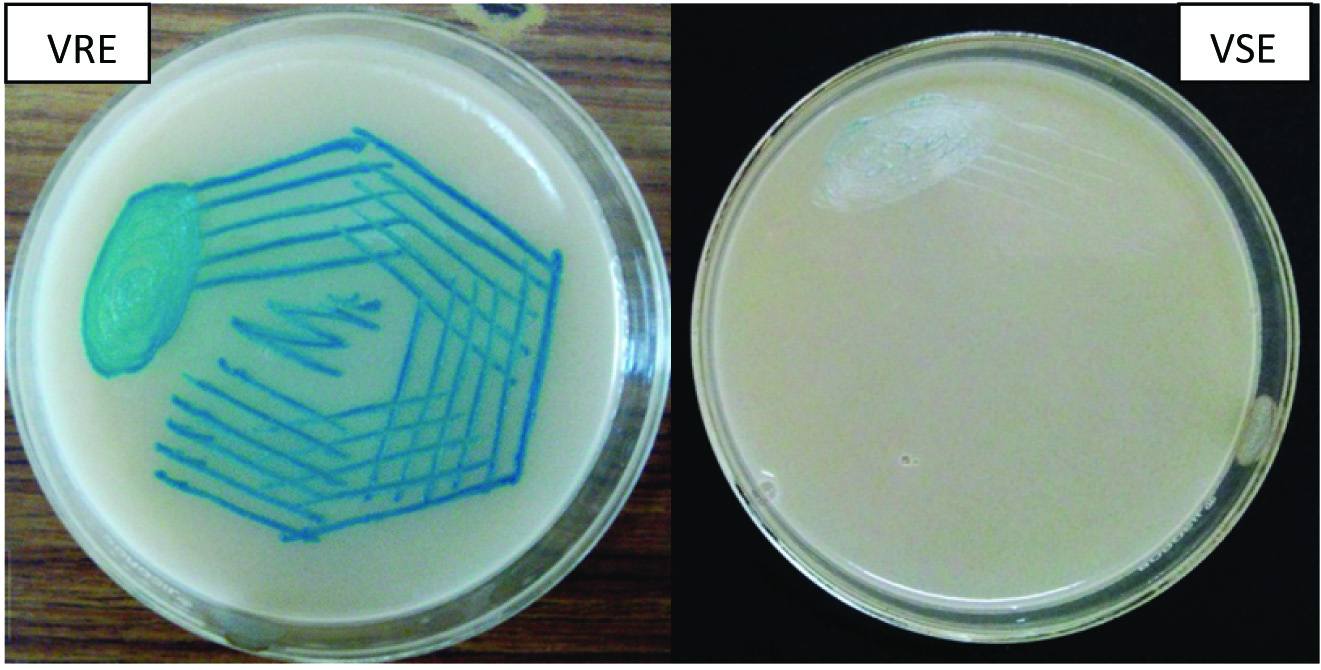
CHROMagarTMVRE and Hicrome VRE were inoculated and incubated aerobically at 37°C. After 48 h of incubation, growth of Enterococci on these chromogenic media indicates VRE. On CHROMagarTMVRE, used for the detection of Van A and Van B type transmissible resistance; vancomycin resistant E.faecalis/E.faecium produce pink to mauve coloured colonies, E.gallinarum and E.casseliflavus resistant to vancomycin produced blue coloured colonies and other Enterococci were inhibited [Table/Fig-2]. On Hicrome VRE agar, vancomycin resistant E.faecalis produced bluish green colonies and others were inhibited [Table/Fig-3].
Vancomycin E-test showing MIC of Enterococci
CHROMagarTM showing VRE positive control and negative isolate
Hicrome VRE showing VRE positive control and negative isolate.
E.faecalis ATCC 29212 and E.faecalis ATCC 51299 were used as susceptible and resistant control strains respectively. Identification of VRE by E- test was considered as reference method.
Results
Out of 4489 clinical samples studied, 100 (2.2%) Enterococci were isolated. [Table/Fig-4]: shows the prevalence of Enterococci in relation to age and sex. [Table/Fig-5]: shows the distribution of Enterococcus species among the various clinical samples. [Table/Fig-6]: shows the antibiotic susceptibility pattern of Enterococci by KBDDM (%). [Table/Fig-7]: shows the Vancomycin MIC range of Enterococci. [Table/Fig-8]: shows the analysis of Chromogenic media with E-test. [Table/Fig-9]: Shows study of VRE as reported by various workers.
Prevalence of Enterococci in relation to age and sex
| Age Years | No of Enteroocci | Male | Female |
|---|
| 0-20 | 10 | 7 | 3 |
| 21-40 | 44 | 15 | 29 |
| 41-60 | 29 | 12 | 17 |
| ≥ 61 | 17 | 9 | 8 |
| Total | 100 | 43 | 57 |
Enterococci species isolated in relation to various clinical samples
| Specimen | Total no | E.faecalis | E.faecium | E.gallinarum | Enterococci (%) |
|---|
| Urine | 1682 | 31 | 28 | 4 | 63(3.75) |
| Exudates | 716 | 15 | 7 | 0 | 22(3.07) |
| Sputum | 434 | 3 | 2 | 1 | 6(1.39) |
| Blood | 943 | 1 | 3 | 0 | 4(0.42) |
| Vaginal swab | 570 | 2 | 1 | 0 | 3(1.05) |
| Body fluids | 144 | 1 | 1 | 0 | 2(1.39) |
| Total | 4489 | 53 | 42 | 5 | 100(2.2) |
Antibiotic susceptibility pattern of Enterococci by KBDDM (%)
| Species | Ampicillin | Penicillin | Ciprofloxacin | Piperacillin | Gentamicin 120 μg | Vancomycin | Lenozolid | Teicoplanin |
|---|
| E.faecalis No.53 | 64.2 | 39.4 | 22.64 | 45.28 | 41.51 | 79.25 | 100 | 66.04 |
| E.faecium No.42 | 52.4 | 23.8 | 23.8 | 40.48 | 47.62 | 11.43 | 100 | 59.52 |
| E.gallinarum No.05 | 100 | 80 | 00 | 60 | 60 | 40 | 100 | 80 |
| 100 | 61 | 35 | 22 | 44 | 45 | 74 | 100 | 64 |
Vancomycin MIC range of Enterococci isolates.
| MIC range μg/ml | E.faecalis | E.faecium | E.gallinarum | Total (%) |
|---|
| ≤ 1 | 20 | 26 | 2 | 48 |
| >1-2 | 27 | 13 | 3 | 43 |
| >2-4 | 06 | 3 | 0 | 09 |
| >4-256 | 0 | 0 | 0 | 0 |
| Total | 53 | 42 | 05 | 100 |
Analysis of Chromogenic media with E-test
| Test | Positive | True positives | False positives | False negatives | True negatives | Sensitivity % | Specificity% |
|---|
| HiChrome agar | 1 | 0 | 1 | 0 | 99 | 100 | 99 |
| CHRO MagarTM | 1 | 0 | 1 | 0 | 99 | 100 | 99 |
Study of VRE as reported by various workers
| Enterococci studied | VRE % | E.faecalis % | E.faecium % | Others % |
|---|
| Vijaya D | 100 | 0 | 0 | 0 | 0 |
| Padmashini [15] | 43 | 4.6% | 72.8% | 16.3% | 6.9% |
| Vidyalakshmi [16] | 600 | 4 | 0 | 100 | 0 |
| Baragundi Mahesh [11] | 120 | 7.5 | 22.2 | 44.44 | 33.33 |
| Gupta [17] | 100 | 2 | 50 | 50 | 0 |
| Neelam Taneja [3] | 144 | 5.55 | 12.5 | 62.5 | 25 |
| Purva Mathur [2] | 444 | 1.12 | 100 | 0 | 0 |
Discussion
Enterococci have attracted much attention in the recent times due to their increased recognition as a cause of nosocomial “super-infection” in patients receiving antimicrobial agents. Enterococci have clearly emerged as a medically important organism in outbreaks of many nosocomial infections. An organism once considered a harmless commensal residing in the intestine has emerged as a multiple drug resistant, virulent pathogen accounting for more hospital borne infection [9].
Enterococci were isolated in 2.22% of the total specimen screened whereas Sreeja reported 0.23% [10]. In India, incidence of Enterococcal infection is not thoroughly identified. E. faecalis is the most prevalent species cultured from humans accounting for 80-90% of clinical isolates in other studies [11].
In the present study, maximum number of Enterococci were isolated from urine samples (3.75%) which is higher than Taneja (1.5%) [3] and Sreeja (1.58%) [10]. Enterococci were isolated from 3.07% of exudates and 0.42% of blood, whereas Sreeja has a higher rate of isolation from exudates (4.47%) and blood (1.1%) [10].
In the present study, E.faecalis were isolated more (53%), which is in comparison with other studies [3,6,11,12].
In the present study, E. faecalis and E. faecium showed almost similar sensitivity to various antibiotics by KBDDM. Resistance to various antibiotics among clinical strains of Enterococci species is a progressive and widely spreading problem. In the present study 55% of the isolate showed high level gentamycin resistance. Similar finding was found in Goshal, whereas Agarwal has reported 7.8% [12,13]. The higher rate of resistance in the present study is attributed to wider usage of broad spectrum antibiotics as this Hospital being a tertiary care Centre.
In the present study, 100% isolates were sensitive to linezolid is in comparison with the report of Gupta and Padmasini [14,15]. Linezolid was the first oxazolidinone to be available for clinical use in 2000. It has activity against both E. faecium and E. faecalis. Another advantage is that it can be administered both intravenously and orally [5]. The pattern of teicoplanin sensitivity in this study correlated with Gupta by disc diffusion method [14].
Vancomycin showed 26% of resistance by KBDDM as shown in the [Table/Fig-4]. By E-test, all Enterococci were sensitive to Vancomycin with MIC <4μg/ml. This proves the inaccuracy of KBDDM in detecting the susceptibility to vancomycin. Others have reported varying percentage of VRE in their studies which is shown in the [Table/Fig-9] [2,3,14–17].
Risk factor for VRE is from exposure to VRE positive patients and lengthy hospital stay. Organ transplantation and hemodialysis patients form the high risk groups, mostly by stool of patient contaminating the environment. Outbreak of VRE can occur from fabric sheets and transferred by staffs’ hands. Vancomycin resistant Enterococci have been shown to be capable of surviving on dry surfaces in the hospital for upto four months [18].
In the present study, CHROMagarTM VRE and Hicrome VRE showed sensitivity of 100% and specificity of 99%, whereas, Llacsahuanga reported sensitivity of 98.2% and specificity of 96.5% and Hajia reported 100% sensitivity and specificity for CHROMagarTMVRE correlating with the present study [7,19]. Jenkins showed sensitivity and specificity of 98% and 95% respectively using a different Chromogenic media [20].
Conventional E-test relies on isolation of the organisms as a first step, then identification of its resistance to the vancomycin on 3rd or 4th day. Therefore, rapid, sensitive and inexpensive methods for detection of VRE are needed. Chromogenic media incorporating Chromogenic enzyme substrates and antimicrobial agents have become available for detection of VRE. E-test cannot be performed directly on clinical specimens, whereas Chromogenic media can be used. Another advantage of CHROMagar, is it can be used for routine screening and identification of VRE in hospitalized patients, thereby routine surveillance will prevent the spread of VRE among patients [7].
Advantage of chromogenic media is that it is rapid, simple, easy to perform, cost effective compared to time consuming, laborious and technically demanding conventional method.
Conclusion
Enterococcus infection should be of concern for health care institution. The early detection of VRE will help in the effective therapy and infection control measures, to prevent the spread of VRE. Chromogenic media has higher sensitivity and specificity in the detection of VRE and can be incorporated in the routine screening. The present study indicates that Chrom agar (Hicrome & CHROMagarTMVRE) in detection of VRE is simple, rapid, easy to perform and cost-effective compared to conventional E- test.