Introduction
Salmonella enterica subspecies enterica serotype Typhi is known as the most common cause of enteric fever, though serotype ParaTyphi A is also implicated sometime. Interestingly, a multi-centric study conducted in Asian countries i.e. China, Pakistan, India, Vietnam, Indonesia [1] along with many other isolated reports from India [2-7] and Nepal [8-9] have revealed that S. ParaTyphi A might be contributing as much as 50% of all the enteric fever cases. Therefore, S. ParaTyphi A has recently been projected as an emerging pathogen.
Emergence of S. ParaTyphi A has got many implications especially for prevention and cure because of: firstly, the approved typhoid fever vaccines (Vi Polysaccharide and live oral Ty21a) do not provide protection against S. ParaTyphi A infection [1]; secondly, if transmission and risk factors for both the serotypes are different then strategies to contain them together should be planned carefully [10]; thirdly, if co-infection with different Salmonella serotypes with different antibiotic sensitivity is present then use of single antibiotic may eradicate one serotype while the other if resistant to that drug may persist. The possibility of co-infection has already been suggested by the population based serological study from Nigeria [4]and nucleic acid based detection in blood from Pakistan in acute enteric fever cases [11]. Moreover, S. ParaTyphi A causes indistinguishable clinical features [8] and may be associated with more complications [12-13]. It is now established that in biological specimens, amplification of specific DNA sequences by nested PCR is better tool than single round PCR. Earlier reports have shown efficiency of this molecular method for detection of Salmonella Typhi in blood samples collected from enteric fever patients [14-15]. However, there is no report where both of the above serotypes have been screened in the same biological samples using nested PCR.
Therefore, we decided to detect Typhi and ParaTyphi A serotypes by nested PCR targeting gene sequences of S. Typhi and S. ParaTyphi A in the stool specimens of healthy control, chronic typhoid carriers and in blood, stool and urine specimens collected from acute cases of enteric fever patients.
Materials and Methods
Ethical consideration
The study plan was approved by Institute Ethics Committee of Banaras Hindu University, Varanasi and informed written consent was obtained from each of the participants/guardians.
Study population
This study was carried out during February, 2010 to July, 2011 in the Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, Varanasi, India. A total of 231 individuals were included in the present study, comprising of 75 cases of afebrile healthy individuals having no history of enteric fever for last one year (group A), 46 cases of chronic typhoid carriers (group B) provisionally diagnosed by physicians on the basis of Widal titres and Indirect Haemagglutination Assay (IHA) and 110 clinically diagnosed enteric fever patients (group C) of these 37 stool and 29 urine specimens could be collected from these 110 enteric fever cases.
Collection and processing of the specimens
About 10 ml of blood was collected by venipuncture aseptically from 110 patients for culture and PCR study, 5 ml of blood were inoculated for bacterial isolation and 5 ml was allowed to clot. The serum was subjected to Widal test and clot was subjected to DNA extraction for PCR analysis. Approximately 10-12 g of stool was collected in 50 ml sterile wide mouth container from each of the 75 afebrile healthy controls, 46 suspected cases of chronic typhoid carriers and 37 from acute typhoid patients. About 40-50 ml of urine was collected in a sterile container from 29 patients.
Serological Analysis
Widal test
Widal test was performed using standard protocol given by manufacturers’ guideline (Span Diagnostics, Surat, India). The antibodies titre 1:160 against either of TO and/or TH and/or AH was considered significant in the present study.
Indirect haemagglutination assay (IHA)
An indirect hemagglutination assay (IHA) measuring antibodies to highly purified Vi polysaccharide antigen was performed according to the method of Barrett (1985) [16]. Glutaraldehyde-treated sheep erythrocytes were sensitized with highly purified Vi antigen (10μl/ml). Serial two fold dilutions of serum samples from 1/40 to 1/320 were added to equal volumes of sensitized cells. The agglutination patterns were read after 2 h of incubation at room temperature and again after overnight incubation at 4°C and titre of ≥160 were considered as significant to diagnose chronic typhoid carriers.
Culture study
About one loop of freshly passed stool sample was directly inoculated on Deoxycholate agar (DCA) and MacConkey agar (MA) plates and about 5 gm of stool specimens was inoculated in selenite-F broth (50 ml) for enrichment of bacteria, and subculture was made on DCA and MA plates after overnight incubation at 37oC. About 5 ml of fresh blood was inoculated in 30 ml bottle containing 0.3% sodium polyanethol sulfonate (SPS) of Brain heart infusion (BHI) broth and incubated at 37oC for bacterial growth. After overnight incubation, subcultures were made on MacConkey agar (MA) and blood agar (BA), plates were incubated overnight at 37oC for bacterial growth. The negative culture bottle were kept in the incubator for one week (7 days), the subculture were made every alternate day on the above mentioned plates. About 15 ml urine was centrifuged for 10000 rpm for 10 min at 4oC, pellet was collected and made suspension in normal saline, half of the suspension was inoculate in selenite-F broth and half of the suspension was spread on DCA and MA with spreader. Incubate both the plates and selenite-F broth for further processing.
Molecular Study
DNA extraction from clinical specimens
Extraction of genomic DNA from freshly collected blood and urine samples was carried out by phenol-chloroform and proteinase-K methods [17]. Three millilitre of freshly collected blood was allowed to clot which was subjected to DNA isolation. Fifteen millilitre of urine was centrifuged at 10000 rpm for 10 min at 4oC. The pellet was subjected for DNA extraction. About 3 g of stool sample was mixed in normal saline and centrifuged at 3000 rpm for 5 min at 4oC to remove debris and supernatant was again centrifuged at 10000 rpm for 10 min at 4oC. Pellet was collected for DNA isolation using the method Van Zwet et al., [18] with slight modifications to minimize PCR inhibitors.
PCR primers
For detection of S. Typhi and S. ParaTyphi A in all the three clinical specimens, nested PCR was used. Oligonucleotide primers were synthesized from the sequence of putative fimbrial protein (stkG) gene sequence of S. ParaTyphi A, (Accession No. CP000026; GI: 56126533). Oligonucleotides stkG F1 and stkG R1 were used in the first round PCR to amplify a 427 bp fragment which correspond to nucleotides 96-118 and 522-501, respectively and for nested PCR, oligonucleotide stkG F2 and stkG R2 were used from amplified product of first round PCR to amplify a 229 bp, correspond to 138-159 and 366-343 respectively of putative fimbrial protein (stkG) gene of S. ParaTyphi A. For the detection of S. Typhi, primers were used for targeting flagellin (fliC) gene sequence described by Song et al.,[19] which was further modified by Frankel [20] and putative fimbrial (staA) gene sequence was also used for detection of S. Typhi described by Pratap et al., [21]. The first round PCR of both genes were amplified 495 and 537 bp where as nested PCR amplified 364 and 377 bp respectively from amplified product of first round PCR [Table/Fig-1].
PCR assay
To check the specificity of the in house designed primers, DNA from different Salmonella serotypes and other organisms were isolated. To find out the minimum detectable level (sensitivity), amplification of the serially diluted DNA isolated from S. ParaTyphi A (ATCC 9150) was subjected for amplification. Further, to evaluate the influence of human DNA on the sensitivity of PCR in different clinical specimens, known amount of DNA (100 ng) from mononuclear cells was spiked to serially diluted DNA from S. ParaTyphi A (ATCC 9150) [Table/Fig-2]. After, establishing the specificity and sensitivity of the newly designed primes performed on DNA isolated from different clinical specimens.
The reaction mixture for the first-round PCR contained 2.5 μl of 10x PCR buffer (100 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl) (Genei, Bangalore, India), 10 pmol of each primer stkG F1 and stkG R1 (SBS Genetech Co., Ltd. Mainland, China), 2 μl (2.5 mM each) of dNTPs mix (Genei, Bangalore, India), 0.33 μl (1 units) of Taq DNA polymerase (Genei, Bangalore, India), 5 μl of DNA template (100 ng), and final volume of 25 μl was adjusted with sterile double distilled water. The amplification reaction was performed in a thermal cycler (Biometra, Goettingen, Germany) with following temperature and duration profile: initial denaturation at 94oC for 5 min followed by 35 cycles for 1 min and denaturation at 94°C, 1 min annealing at 57°C, and 1 min extension at 72°C, with a final extension at 7 min. The nested PCR master mix was the same as that of the first round PCR, except it contained 10 pmol of each primer stkG F2 and stkG R2 and 2 μl of DNA template (amplified product of the primary cycle). Thermal cycling was carried out as described for first-round PCR, except that the annealing temperature was set to 61°C. The amplification was repeated 2-3 times to ensure that the amplification obtained with the primers is reproducible and consistent.
Ten millilitres of amplicon was subjected to electrophoresis on agarose gel (1.5% w/v) in TBE buffer (Tris-borate-EDTA) by adding 2 μl ethidium-bromide (10 mg/ml) at 85 V for 1 h and bands were visualized in a gel documentation system (Alfa Imager 2200, Alfa Innotech Corporation, California, USA). Although, all the essential precautions were taken to avoid laboratory contamination, known positive (DNA from reference strain of S. Typhi, MTCC 3216) and negative controls (distilled water) were put to rule out the contamination in each set up.
Restriction digestion
The sequence of fimbrial protein (stkG) gene of S. ParaTyphi A (ATCC 9150) were accessed from the National Centre for Biotechnology Information (NCBI) Gene Bank and subjected to in silico HaeIII restriction digestion by using NEB Cutter version 2.0. The actual amplicons of S. ParaTyphi A were suggested to the enzyme and compared with the in silico results.
Statical Analysis
To test the level of significance between two proportions i.e. nested PCR positivity among group A, group B and group C, by using student t-test.
Results
Serological analysis
In the acute typhoid cases, Widal test, all the 110 (100%) sera from suspected cases had anti TO titre ≥1:160 while 61.8% (68/110) and 38.2% (42/110) had raised titre against TO/TH and AH or both antibodies at titre ≥1:160 (group C) [Table/Fig-3]. However, a total of 41.3% (19/46) (group B) patients were found to be positive for ViAb at a significantly high titre i.e. ≥160 by indirect haemagglutination assay (IHA).
Culture isolation and identification of S. Typhi and ParaTyphi A from blood and stool specimens
A total of 1.33% was observed culture positive in stool specimens of healthy individuals (group A), 4.34% in suspected chronic typhoid carriers (group B) and 19% in acute typhoid cases (group C) by conventional method. Moreover, the total positivity of both serotypes were found to be 30.9% (34/110) from blood specimens of acute typhoid cases (group C), of these we could observe 28.2% (31/110) for S. Typhi as well as 2.7% (3/110) for S. ParaTyphi A but none of the urine samples yielded growth for both serotypes [Table/Fig-4&Table/Fig-5].
The isolated strains were confirmed by Gram’s staining, biochemical test and suspected colony was further confirmed by serological agglutination using poly O, poly H, factors O9, H-d and Vi antisera for S. Typhi and factor-a (H-a) for S. ParaTyphi A. The isolates were Gram negative, rod shape and gives standard biochemical reaction with both serotypes.
Specificity of the primers
Other Salmonella serotypes and other enterobacteriaceae group of bacteria could not yield the desired amplicon i.e. 427 bp and 229 bp by first and second round of PCR while the desired amplicons were obtained from the DNA of S. ParaTyphi A only [Table/Fig-2].
Sensitivity of the primers
First round of PCR only 3 ng of S. ParaTyphi A DNA (corresponding to 3x104 CFU/ml) produced an amplification product of 427 bp on agarose gel in spiked specimens. The nested PCR could amplify the desired size of amplicon 3 fg of target DNA (corresponding to 1 CFU/ml) [Table/Fig-6,7].
Restriction analysis of fimbrial protein (stkG) gene sequence
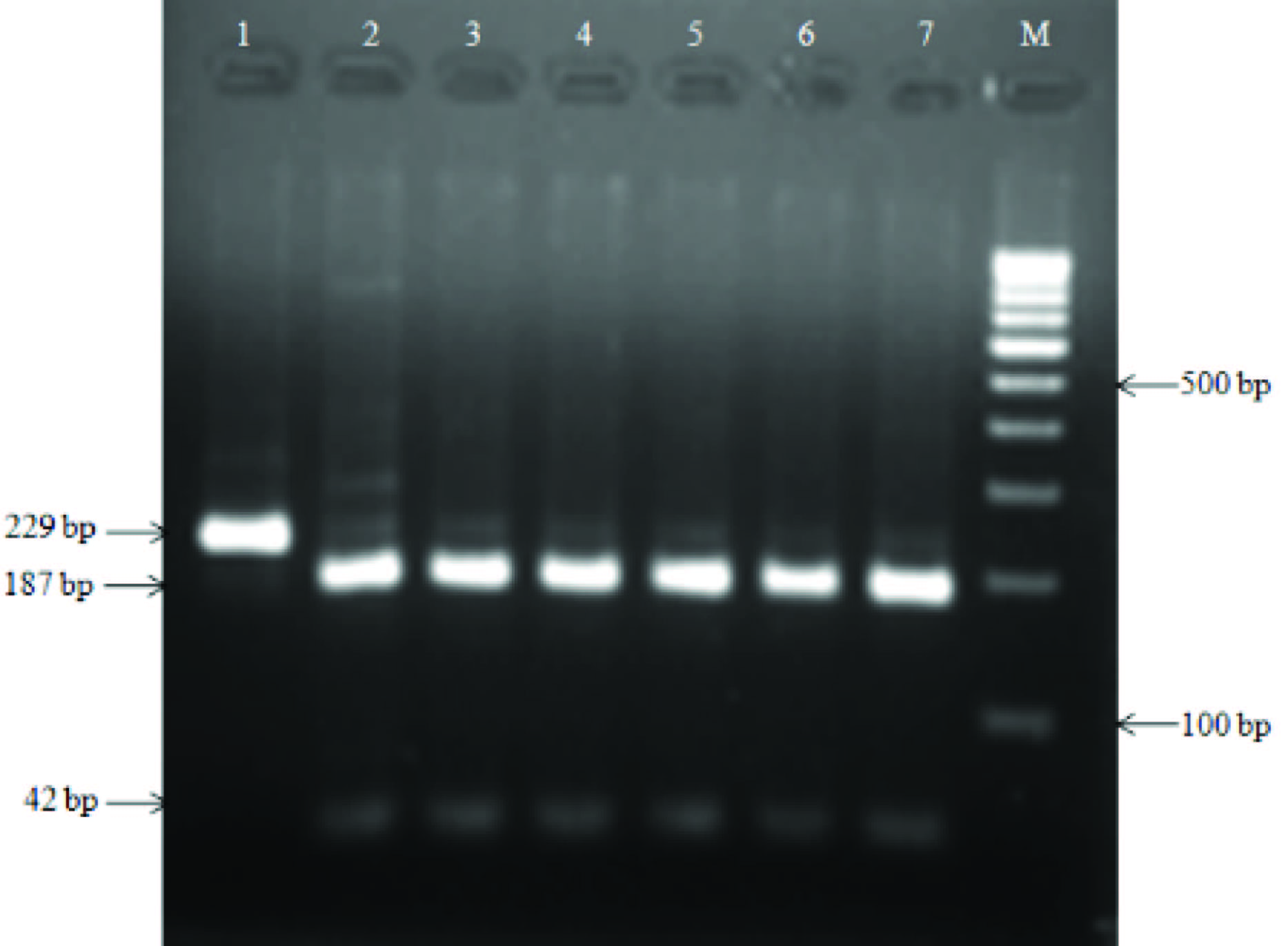
The 229 bp sized amplicon could be restricted into two bands of 187 bp and 42 bp by HaeIII restriction enzyme. The restriction patterns were identical to that of in silico generated bands from S. ParaTyphi A (ATCC 9150) stkG gene sequences [Table/Fig-8]
Amplification of S. Typhi and S. ParaTyphi A in healthy individuals
Of the 75 stool samples from healthy controls, 17.3% (13/75) were positive for both the serotypes Typhi and ParaTyphi A, however 2.7% (2/75) of the specimens were positive for S. Typhi and 1.33% (1/75) for S. ParaTyphi A exclusively. On the other hand, overall S. ParaTyphi A could be detected 14.7% (11/75) of the stool specimens [Table/Fig-4].
Amplification of S. Typhi and S. ParaTyphi A in chronic typhoid carriers
We observed a total of 38/46 (82.6%) of the specimens were positive for either or both Salmonella serotypes. Compared to healthy controls, for which we observed 13/75 (17.3%) positivity, the chronic typhoid carriers have a significantly higher (p<0.001) rate of positivity for the Salmonella serotypes [Table/Fig-4].
Amplification of S. Typhi and S. ParaTyphi A in clinical specimens of typhoid cases
The desired amplicons were observed 92.7% (102/110) for S. Typhi and 18.2% (20/110) for ParaTyphi A in blood specimens. The total positivity for S. Typhi and S. ParaTyphi A altogether reached 94.5% (104/110). Co-infection could be observed in 16.4% (18/110) while exclusive positivity for S. Typhi and ParaTyphi A were in 76.4% (84/110) and 1.81% (2/110) respectively.
Of the 37 stool specimens, 59.5% (22/37) were found to be positive for S. Typhi while 32.4% (12/37) for S. ParaTyphi A. Consequently, 78.4% of the total samples were positive for Salmonella serotypes shows statistically significant (p<0.001) (group A vs group C). Five (13.5%) stool specimens had co-infection while 45.9% (17/37) patients had S. Typhi and 18.9% (7/37) S. ParaTyphi A exclusively in their stool samples [Table/Fig-4].
Of the total 29 urine samples the detection rate reached 100% (29/29) (p<0.001) when both Salmonella serotypes were taken together. S. Typhi could be detected in 93.1% (27/29) while S. ParaTyphi A in 41.4% (12/29). Co-infection could be detected in 34.4% (10/29) of the specimens while S. Typhi and S. ParaTyphi A could be detected in exclusively 58.6% (17/29) and 6.9% (2/29) of the urine specimens respectively [Table/Fig-5,9].
Discussion
In this study, detection of both the serotypes (Typhi and ParaTyphi A) in >40% of the urine samples collected from enteric fever cases by nested PCR indicates that co-infection by the two serotypes is quite frequent phenomenon. Interestingly, when we carried out the detection of S. Typhi and ParaTyphi serotypes by nested PCR in stool specimens of healthy controls and clinically suspected cases of typhoid carriers the positivity rate for S. ParaTyphi was quite high in the later group (69.6%) while it was 14.7% in healthy controls (14.7%). These findings suggest that S. ParaTyphi A infection is not infrequent in the north India at least. However, it is really difficult to explain the positivity of S. ParaTyphi A in chronic carriers at such a high rate. However, several speculations can be made e.g. the chronic carriers are more susceptible to Salmonella infection and their immune system is unable to eradicate both S. Typhi as well as ParaTyphi A. Although clinically the acute infection caused by S. ParaTyphi A and Typhi cannot be differentiated bit it is believed that ParaTyphi A causes milder disease and thus ParaTyphi A is quite frequent in chronic carriers. However, further studies are needed to explore the causation of such a high occurrence of S. ParaTyphi A in chronic carriers. These findings suggest that S. ParaTyphi A is not an emerging infection but it is naturally prevalent in the typhoid endemic areas. In support of this statement, a study from Nigeria, has reported that 15.6% of the enteric fever patients had co-infection as titre were found raised against both the flagellar antigens i.e. TH and AH [22]. The other study, based on nucleic acid amplification from blood samples of enteric fever patient’s has also shown 12% positivity for both the serotypes[11]. Interestingly, 16.4% of the blood samples in the present study were also found positive for the mixed infection. There are a few case reports from Indian subcontinent showing isolation of both the serotypes from enteric fever patients [23-25]. A study conducted in four countries of Asia during 2001-2003 has reported percentage positivity for S. ParaTyphi A to be 15% in Pakistan, 24% in India, 14% in Indonesia and 64% in China [1]. Do the above reports indicate real increase in the incidence of S. ParaTyphi A and decline of S. Typhi? The possible explanation for this observation may be that: incidence of S. Typhi might be decreasing but increasing for S. ParaTyphi A or it is because of simply screening the bacterial colonies producing gas along with H2S with ParaTyphi A specific antisera as well since both the serotypes are growing on the culture media or it may be due to increased awareness for the detection of S. ParaTyphi A due to certain outbreaks by this serotypes. Our study is unique in the sense that we have applied very sensitive and specific detection method and observing co-infection at such a high level. Here, it will be worth mentioning that putative fimbrial protein (stkG) gene also shares with S. Heidelberg, Bareilly, and Thompson serotype, but this serotypes usually does not causes enteric fever as well as the prevalence might not be quite common in this region but it is further required to explore the actual prevalence of S. ParaTyphi A targeting some other specific gene sequences. Although, there is a report suggesting that outbreaks of S. ParaTyphi A may occur due to acquisition of drug resistance and other virulence factors by the serotype providing better opportunity for its survival[26], our study expanded over a period of one year and patients were from wide spread area (100 km radius), the possibility of outbreak seems to be unlikely. Therefore, the reported rise in the incidence of S. ParaTyphi A may not be real and the co-infection by S. Typhi and S. ParaTyphi A might be quite frequent occurrence. Therefore, it may be suggested that all the cases of enteric fever must be screened for the presence of both the serotypes in an enteric fever patient.
The co-infection observed in the present study is quite likely as the source and routes of transmission are similar. On the basis of this study, it may be suggested that the similar looking colonies on the culture plates might be of different serotypes of Salmonella implicated in enteric fever and several of these colonies may be screened by specific antisera. While similar treatment strategies mostly work for both organisms, the two serotypes with different antibiograms may affect the outcome of antibiotic therapy in the era of emergence of multi drug-resistant strains. Further, future enteric fever prevention strategies in Asia must focus on S. ParaTyphi A as well, Bivalent vaccines that protect against S. Typhi as well as S. ParaTyphi A seems to be the need of the hour as the protective efficacy of typhoid fever vaccines (Vi, Ty21a) focussing S. Typhi only may diminish resulting into loss of public confidence and decrease public willingness to participate in mass vaccination programme.
Primers used for detection of S. Typhi and S. ParaTyphi A serotypes in clinical specimens., PCR: Polymerase chain reaction
| Gene | Primer | Primer sequence (5’-3’) | Products size | Target positions | PCR condition, (number of cycles) | References |
| Flagellin (fliC) gene |
| Primary | ST1 | ACTGCTAAAACCACTACT | 495 bp | 1036-1056 | 94oC, 5 min; 52oC, 1 min; 72oC, 1 min, (35) | [19,20] |
| ST2 | TTAACGCAGTAAAGACAG | | 1513-1530 | | |
| Nested | ST3 | AGATGGTACTGGCGTTGCTC | 364 bp | 1072-1089 | 94oC, 5 min; 63oC, 1 min; 72oC, 1 min, (35) | |
| ST4 | TGGAGACTTCGGTCGCGTAG | | 1416-1435 | | |
| Putative fimbrial (staA) gene |
| staA F1 | TGGTTACATGACCGGTAGTC | 537 bp | 33-52 | 94oC, 1 min; 56oC, 1min; 72oC, 1 min (35) | [21] |
| staA R1 | TAGCTGCCGCAATGGTTATG | | 569-550 | | |
| staA F2 | CATCGGCACGAACGTAAGAC | 377 bp | 66-85 | 94oC, 1 min; 63oC, 1min; 72oC, 1min (35) | |
| staA R2 | TCAAGCGACTGATGGTGACG | | 442-423 | | |
| Putative fimbrial protein (stkG) gene |
| Primary | stkG F1 | CGTTTACTGAGGTCACAGGCATC | 427 bp | 96-118 | 94oC, 5 min; 57oC, 1 min; 72oC, 1 min, (35) | This study |
| stkG R1 | CACATTGTTCTCGGAGACCCCA | | 522-501 | | |
| Nested | stkG F2 | CAATGGCTTCTGGCGAACTGTC | 299 bp | 138-159 | 94oC, 5 min; 61oC, 1 min; 72oC, 1 min, (35) | |
| stkG R2 | GTGGAGAAAGATCAGACCACCGAG | | 366-343 | | |
List of bacteria subjected for testing the specificity of stkG gene specific primers
| Salmonella enterica subspecies enterica reference strains | No. of isolates | Antigen structure | stkG gene |
|---|
| Sero-group | Serovar | O | H-1 | H2 |
|---|
| A | ParaTyphi A (ATCC 9150) | 1 | 1,2,12 | a | - | + |
| ParaTyphi B (ATCC 10719) | 1 | 4,5,12 | b | 1,2 | - |
| B. | Typhimurium (ATCC700720) | 1 | 4,5,12 | i | 1,2 | - |
| Heidelberg (ATCC 41578) | 1 | 1 ,4,5,12 | r | 1,2 | + |
| C. | ParaTyphi C (ATCC 13428) | 1 | 6,7 | c | 1,5 | - |
| D. | Typhi (MTCC 3216) | 1 | 9,12,(Vi) | d | - | - |
| Enteritidis (ATCC 13076) | 1 | 1,9,12 | g, m | - | - |
| Clinical isolates (Salmonella and other gram negative bacteria) |
| Salmonella ParaTyphi A | 27 | + |
| Salmonella Typhimurium | 15 | - |
| Salmonella Typhi | 36 | - |
| Salmonella ParaTyphi C | 5 | - |
| Salmonella ParaTyphi B | 4 | - |
| Citrobacter freundii | 1 | - |
| Pseudomonas aeruginosa | 3 | - |
| Shigella dysenteriae | 5 | - |
| Shigella flexneri | 6 | - |
| Proteus vulgaris | 3 | - |
| Proteus mirabilis | 4 | - |
| Escherichia coli | 1 | - |
| Klebsiella pneumonia | 3 | - |
| Morganella morganii | 5 | - |
Showing comparative analysis of Widal and PCR test carried out in the peripheral blood specimens (n=110)
| Widal test (at titre >1:160 either TO or TH or AH or in combination) | Nested PCR |
|---|
| No. of positive | Percentage positivity (%) | No. of positive | Percentage positivity (%) |
|---|
| 110 | 100 | 104 | 94.5 |
Culture isolation and PCR positivity of S. Typhi and ParaTyphi A in all the available stool specimens in three different groups.
| Clinical specimens (used for culture isolation and nested PCR) | Culture positivity (%) | PCR positivity (%) |
|---|
| S. Typhi alone (%) | S. ParaTyphi A alone (%) | S. Typhi + S. ParaTyphi A (%) | Total S. Typhi (%) | S. ParaTyphi A (%) | Total (%) |
|---|
| Group A (n=75) (age group 2-60 years) | 1 (1.33) | 2 (2.7) | 1 (1.3) | 10 (13.3) | 12 (16.0) | 11 (14.7) | 13 (17.3) |
| Group B (n=46) (age group 18-60 years) | 2 (4.34) | 6 (13.0) | 4 (8.7) | 28 (60.8) | 34 (73.9) | 32 (69.6) | 38 (82.6) |
| Group C (n=37) (age group 3-42 years) | 7 (19) | 17 (45.9) | 7 (18.9) | 5 (13.5) | 22 (59.5) | 12 (32.4) | 29 (78.4) |
1P-value <0.001 between Group A vs Group B, Group A vs Group C, but 1P-value no significant (NS) between Group B vs Group C, (total S. Typhi (alone + Mix infection); 2P <0.050 between Group A vs Group B, Group A vs Group C and Group B vs Group C, (total S. ParaTyphi A (alone + Mix infection); 3P-value <0.001 between grant total of Group A vs Group B, Group A vs Group C, but 3P-value no significant (NS) between Group B vs Group C; NS-no significance P-value
Detection of S. Typhi and S. ParaTyphi A in clinical specimens.
| Clinical specimens (used for culture and nested PCR) Group C (age group 3-42 years) | Culture positivity (%) | Nested PCR positivity (%) |
|---|
| S. Typhi (%) | S. ParaTyphi A (%) | Total (%) | S. Typhi alone (%) | S. ParaTyphi A alone (%) | S. Typhi + S. ParaTyphi A (%) | Total S. Typhi (%) | Total S. ParaTyphi A (%) | Total (%) |
|---|
| Blood (n=110) | 31 (28.2) | 3 (2.7) | 34 (30.9) | 84 (76.4) | 2 (1.81) | 18 (16.4) | 102 (92.7) | 20 (18.2) | 104 (94.5) |
| Stool (n=37) | 7 (19) | 0 | 7 (19) | 17 (45.9) | 7 (18.9) | 5 (13.5) | 22 (59.45) | 12 (32.43) | 29 (78.4) |
| Urine (n=29) | 0 | 0 | 0 (0) | 17 (58.6) | 2 (6.9) | 10 (34.4) | 27 (93.1) | 12 (41.4) | 29 (100) |
1P-value <0.001 between Blood vs Stool, Blood vs Urine, Stool vs Urine of total S. Typhi (alone + Mix infection); 2P-value <0.050 between Blood vs Stool, Blood vs Urine, Stool vs Urine of total S. ParaTyphi A (alone + Mix infection); 3P-value <0.050 between grant total of Blood vs Stool, Blood vs Urine, Stool vs Urine
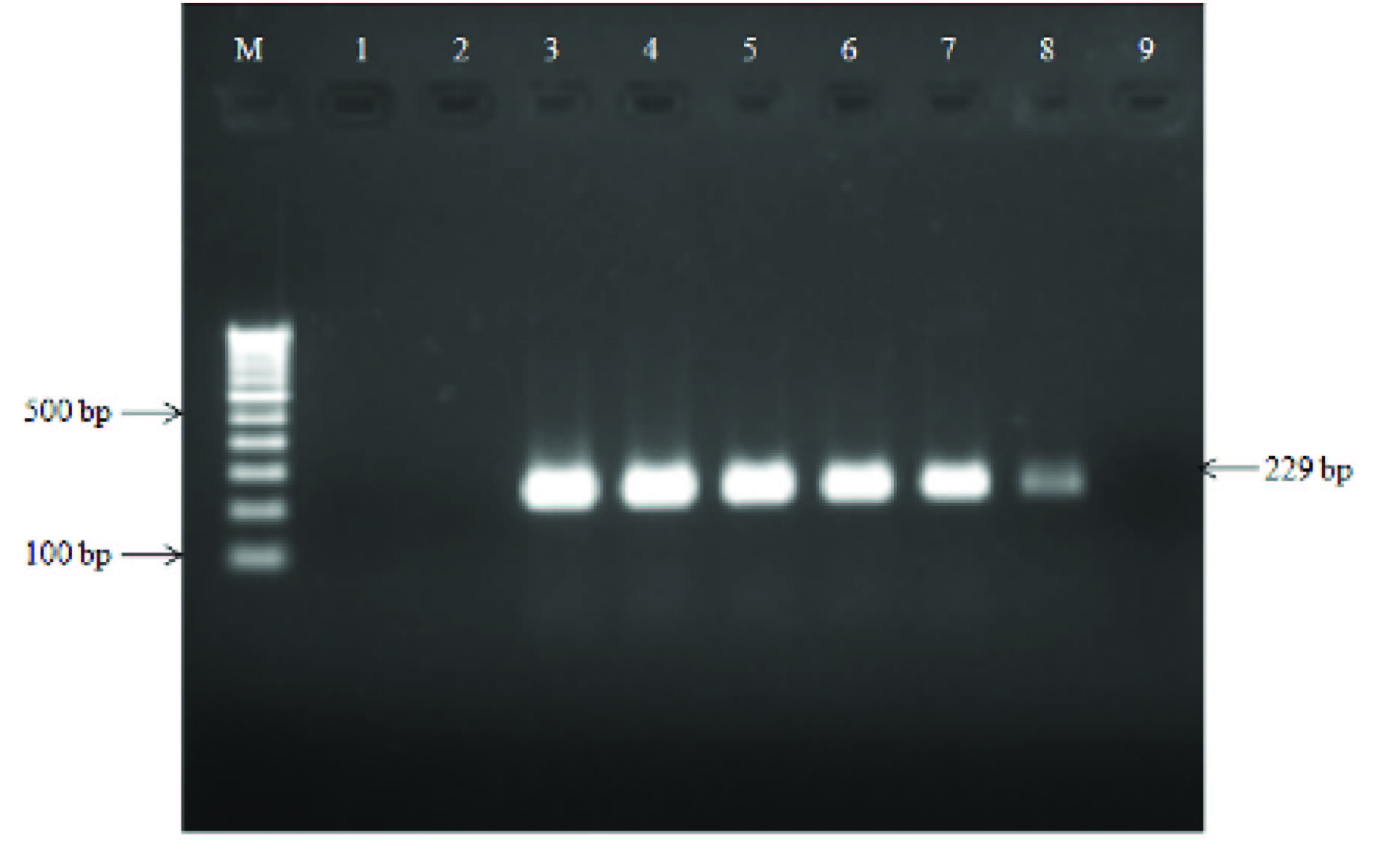
Sensitivity testing of stkG gene primers in serially diluted Salmonella ParaTyphi A genomic DNA; lane M, 100 bp DNA ladder; lane 1, negative control; lane 2, positive control (199 ng/μl); lane 3-10 represent serially diluted S. ParaTyphi A DNA from 100 ng/μl to 0.1 fg/μl
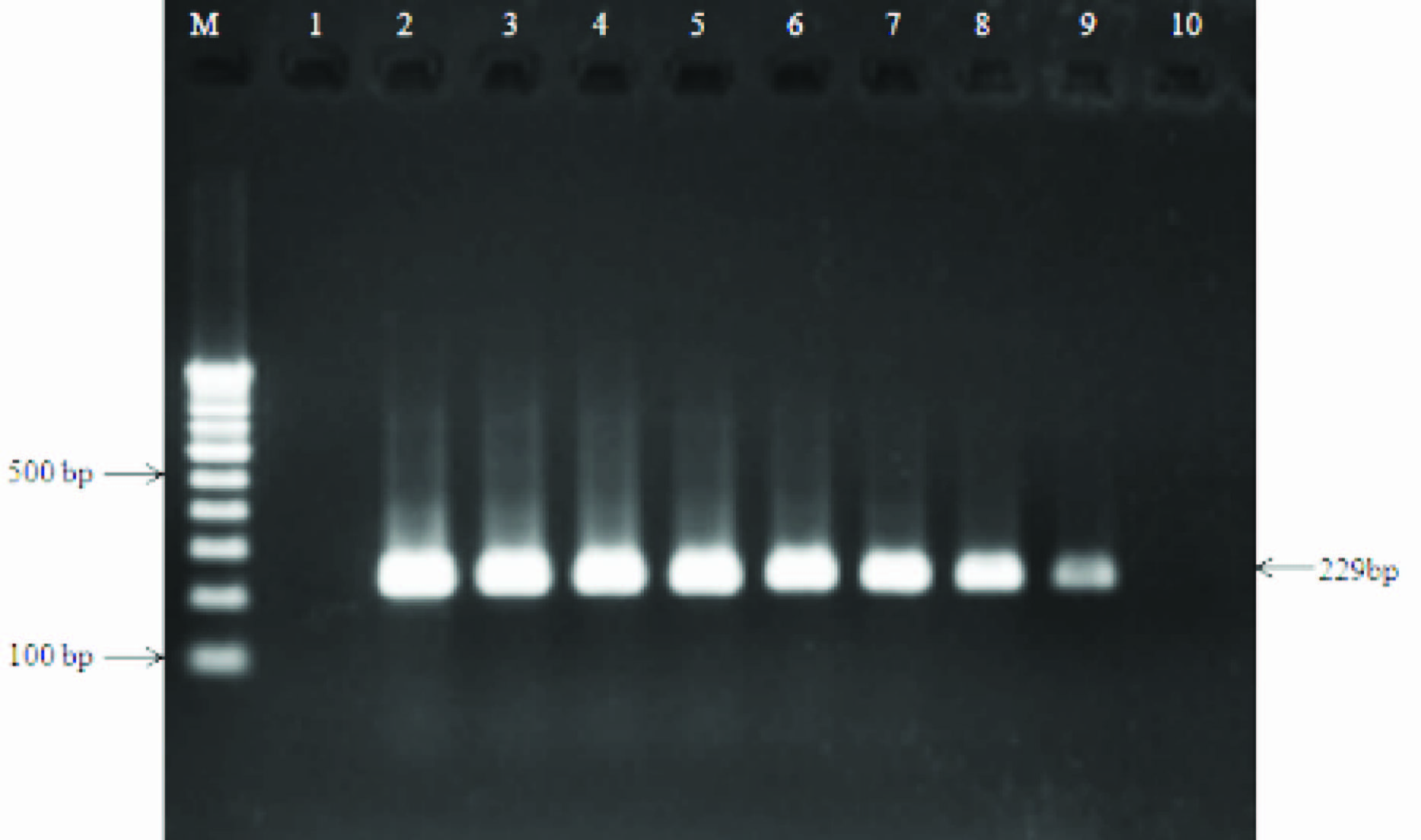
Sensitivity testing of stkG gene primers in spiked human blood with S. ParaTyphi A (ATCC9150) detected 3 CFU/ml. Lane 1, negative control; Lanes 2, control with leukocyte DNA without S. ParaTyphi A DNA; lane 3, 3x108 cell/ml (S. ParaTyphi A ATCC9150), lane 4, 3x106 cell/ml, lane 5, 3x104 cell/ml; lane 6, 3x102 cell/ml; lane 7, 3x101 cell/ml; lane 8, 3x100 cell/ml; lane 9, 1x100 cell/ml
HaeIII restriction digestion of 229 bp amplicon targeting stkG gene; Lane M, 100 bp DNA ladder; lanes 1, unrestricted amplicon of S. ParaTyphi A (ATCC9150), lane 2-7 restricted fragments of stkG gene amplicon of clinical samples
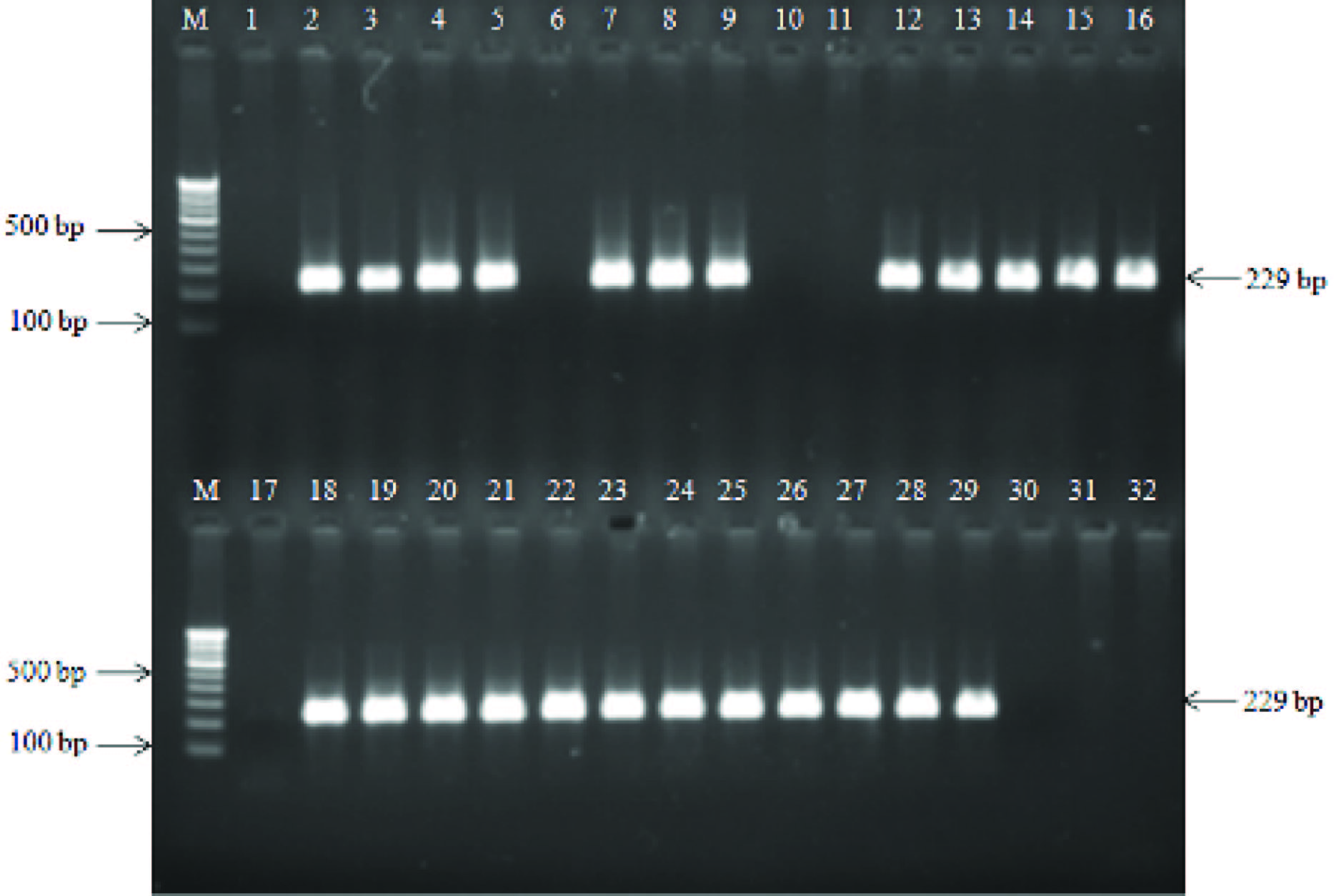
Nested PCR amplification targeting of S. ParaTyphi A specific fimbrial protein (stkG) gene in clinical samples. Lane M, 100 bp DNA ladder; lane 1, negative control; lane 2, positive control (S. ParaTyphi A ATCC9150); lane 6,10,17,30,31 and 32, negative nested PCR amplification; lanes 3-5,7-9,12-16,18-29 positive nested PCR amplification in different clinical samples
Conclusion
Serological confirmation may be done by sweeping the bacterial colonies and if it gives positive agglutination for both the serotypes Typhi and ParaTyphi A, the colonies must be separated and antibiotic susceptibility testing should be done for each of the serotypes and if indicated multidrug therapy may be instituted. Apart from diagnostic and therapeutic implications, we must look for the vaccines capable of providing protection against both the serotypes.
1P-value <0.001 between Group A vs Group B, Group A vs Group C, but 1P-value no significant (NS) between Group B vs Group C, (total
S. Typhi (alone + Mix infection); 2P <0.050 between Group A vs Group B, Group A vs Group C and Group B vs Group C, (total S. ParaTyphi A (alone + Mix infection); 3P-value <0.001 between grant total of Group A vs Group B, Group A vs Group C, but 3P-value no significant (NS) between Group B vs Group C; NS-no significance P-value
1P-value <0.001 between Blood vs Stool, Blood vs Urine, Stool vs Urine of total S. Typhi (alone + Mix infection); 2P-value <0.050 between Blood vs Stool, Blood vs Urine, Stool vs Urine of total S. ParaTyphi A (alone + Mix infection); 3P-value <0.050 between grant total of Blood vs Stool, Blood vs Urine, Stool vs Urine
[1]. RL Ochiai, X Wang, L von Seidlein, J Yang, ZA Bhutta, SK Bhattacharya, Salmonella ParaTyphi A rates, AsiaEmerg Infect Dis 2005 11(11):1764-66. [Google Scholar]
[2]. S Sood, A Kapil, N Dash, BK Das, V Goel, P Seth, Paratyphoid fever in India: an emerging problemEmerg Infect Dis 1999 5(3):483-84. [Google Scholar]
[3]. DS Chandel, R Chaudhry, B Dhawan, A Pandey, AB Dey, Drug resistant Salmonella enterica serotype ParaTyphi A in IndiaEmerg Infect Dis 2000 6(4):420-21. [Google Scholar]
[4]. SS Tankhiwale, G Agrawal, SV Jalgaonkar, An unusually high occurrence of Salmonella enterica serotypes ParaTyphi A in patients with enteric feverIndian J Med Res 2003 117:10-12. [Google Scholar]
[5]. BN Harish, GA Menezes, K Sarangapani, SC Parija, Fluoroquinolone resistance among Salmonella enterica serovar ParaTyphi A in PondicherryIndian J Med Res 2006 124:585-87. [Google Scholar]
[6]. SS Bhattacharya, U Dash, A sudden rise in occurrence of Salmonella ParaTyphi A infection in Rourkela, OrissaIndian J Med Microbiol 2007 25(1):78-79. [Google Scholar]
[7]. V Gupta, J Kaur, J Chander, An increase in enteric fever cases due to Salmonella ParaTyphi A in & around ChandigarhIndian J Med Res 2009 129(1):95-98. [Google Scholar]
[8]. AP Maskey, JN Day, QT Phung, GE Thwaites, JI Campbell, M Zimmerman, Salmonella enterica serovar ParaTyphi A and S. enterica serovar Typhi cause indistinguishable clinical syndromes in Kathmandu, NepalClin Infect Dis 2006 42(9):1247-53. [Google Scholar]
[9]. D Acharya, DR Bhatta, S Malla, SP Dumre, N Adhikari, BP Kandel, Salmonella enterica serovar ParaTyphi A: an emerging cause of febrile illness in NepalNepal Med Coll J 2011 13(2):69-73. [Google Scholar]
[10]. AM Vollaard, S Alim, HA van Astenm, S Widjaja, LG Visser, C Surjadi, Risk factors for typhoid and paratyphoid fever in Jakarta, IndonesiaJAMA 2004 291(21):2607-15. [Google Scholar]
[11]. A Ali, A Haque, A Haque, Y Sarwar, M Mohsin, S Bashir, Multiplex PCR for differential diagnosis of emerging typhoidal pathogens directly from blood samplesEpidemiol Infect 2009 137(1):102-07. [Google Scholar]
[12]. R Chaudhry, RK Mahajan, A Diwan, S Khan, R Singhal, DS Chandel, Unusual presentation of enteric fever: three cases of splenic and liver abscesses due to Salmonella Typhi and Salmonella ParaTyphi ATrop Gastroenterol 2003 24(4):198-99. [Google Scholar]
[13]. P Navin, DS Thambu, R Venugopal, HS Subhash, K Thomas, Salmonella ParaTyphi osteomyelitis and psoas abscessTrop Doct 2006 36(1):58-59. [Google Scholar]
[14]. P Prakash, OP Mishra, AK Singh, AK Gulati, G Nath, Evaluation of Nested PCR in Diagnosis of Typhoid FeverJ Clin Microbiol 2005 43(1):431-32. [Google Scholar]
[15]. G Kumar, CB Pratap, OP Mishra, K Kumar, G Nath, Urine in the diagnosis of typhoid fever by using nested PCR targeting flagellin gene (fliC)J Clin Microbiol 2012 50(6):1964-67. [Google Scholar]
[16]. TJ Barrett, Improvement of the indirect hemagglutination assay for Salmonella Typhi Vi antibodies by use of glutaraldehyde-fixed erythrocytesJ Clin Microbiol 1985 22:662-63. [Google Scholar]
[17]. J Sambrook, DW Russell, Molecular cloning: a laboratory manualLaboratory Press 2001 3rd EditionCold Spring Harbor, NYCold. Spring. Harbor:1-31. [Google Scholar]
[18]. AA Van Zwet, JC Thijs, AM Kooistra-Smid, J Schirm, JAM Snijder, Use of PCR with faeces for detection of Helicobacter pylori infections in PatientsJ Clin Microbiol 1994 32(5):1346-48. [Google Scholar]
[19]. JH Song, H Cho, MY Park, DS NA, HB Moon, CH Pai, Detection of Salmonella Typhi in the blood of patients with typhoid fever by polymerase chain reactionJ Clin Microbiol 1993 31(6):1439-43. [Google Scholar]
[20]. G Frankel, Detection of Salmonella Typhi by PCRJ Clin Microbiol 1994 32(5):1415 [Google Scholar]
[21]. CB Pratap, G Kumar, SK Patel, AK Verma, VK Shukla, K Kumar, Targeting of putative fimbrial gene for detection of S. Typhi in typhoid fever and chronic typhoid carriers by nested PCRJ Infect Dev Ctries 2013 7(7):520-27. [Google Scholar]
[22]. EU Umeh, CO Agbulu, Distribution Pattern of Salmonella Typhoidal Serotypes in Benue State Central, NigeriaInter J Epidemiol 2010 8(1):3 [Google Scholar]
[23]. JN Devi, PV Rao, PG Shivananda, Enteric fever due to double Salmonella serotypeIndian J Pathol Microbiol 1986 29(2):209-10. [Google Scholar]
[24]. S Joshi, C Wattal, A Sharma, KJ Prasad, Mixed Salmonella infection- a case reportIndian J Med Micribiol 2002 20(2):113-14. [Google Scholar]
[25]. N Perera, C Geary, M Wiselka, K Rajakumar, SR Andrew, Mixed Salmonella infection: Case report and review of the literatureJ Travel Med 2007 14(2):134-35. [Google Scholar]
[26]. O Gal-Mor, J Suez, D Elhadad, S Porwollik, E Leshem, L Valinsky, Molecular and cellular characterization of a Salmonella enterica serovar ParaTyphi A outbreak strain and the human immune response to infectionClin Vaccine Immunol 2012 19(2):146-56. [Google Scholar]