Background:Salmonella, a genus of more than 2500 serological variants (serovars), includes many organisms that can cause human disease. Enteric fever remains an important public health problem in developing countries. Non typhoidal Salmonella generally produce a self limited gastroenteritis in healthy individuals whereas in extremes of age and immunocompromised cause severe fatal disease. The protean manifestations make this disease a true diagnostic challenge.
Aim: The present study was carried out to optimize PCR for detecting the Salmonella genus using iroB gene and evaluate its use in the rapid diagnosis of typhoid and non typhoidal salmonellosis.
Materials and Methods: The study was carried out between August 2009 and July 2011 on blood samples from patients attending JIPMER hospital, Pondicherry, India with clinical suspicion of enteric fever and salmonellosis. Whole blood was used DNA extraction and conventional PCR done with iroB and fliC primers. Blood culture and Widal test were performed for all the patients.
Statistical Analysis: Performed using Fischer’s exact test with Graphpad Instat 3.
Results: PCR results were compared with blood culture. Sensitivity and specificity of PCR with fliC gene are 95.6% and 93.3% respectively. Sensitivity and specificity of PCR with iroB gene are 96.6% and 93.3% respectively
Conclusion: With iroB gene, additional cases of Salmonella Paratyphi A and non typhoidal Salmonella were detected when compared to fliC gene.
Introduction
Salmonellae are facultative intracellular pathogens. Salmonella enterica serovar Typhi is exclusively host-restricted to humans, and early intracellular replication within reticuloendothelial mononuclear cells is critical for pathogenesis [1]. In contrast, non-typhoid Salmonella (NTS) serovars have a broad host range and usually cause self-limiting mucosal diarrheal disease and carriage in immunocompetent humans [2].
Culturing blood specimens has become much faster and easier, since the advent of automated blood culture instruments such as the BACTEC 9000 systems (Becton Dickinson, Cockeysville, MD, USA) and BacTAlert (BioMerieux, Durham, NC, USA). However, it still takes several days to detect and identify Salmonella [3,4]. Due to the time required for blood culture identification alternative methods of identification of Salmonella are being sought. In particular, DNA detection methods such as the polymerase chain reaction (PCR) have been investigated [5]. PCR-based typing techniques are becoming increasingly popular in developing countries. PCR can be useful in salmonellosis especially in cases when empirical antibiotic is started and bacterial load is low.
Many of the molecular methods done for detection of Salmonella in blood were based on using fliC gene which is specific for Salmonella typhi. The flagellum of S. typhi like all other bacterial flagella is Composed of pre-distinct portion, basal body, hook and filament. The filament is made up of repeating subunits of a single protein, flagellin. Salmonella typhi typically has only phase I flagellar antigen, d. This d antigen however is the phase I antigen in a number of Salmonella species like Salmonella muenchen. A unique sequence present in the hypervariable region VI of S. typhi distinguishes it from all other Salmonella spp especially S.muenchen. PCR for S.typhi utilizes primers designed to detect this region of the flagellin gene. Studies have shown a good specificity with these primers [6]. On the other hand, the incidence of enteric fever caused by S. Paratyphi A has been increasing in Asia. It is estimated to have more than 5 million cases of paratyphoid fever compared to more than 25 million cases of typhoid fever [7]. Typhoid is a highly adapted invasive Parija4disease that is restricted to humans and shows little association with the immunocompromised. In contrast, non-typhoidal salmonellosis have a broad vertebrate host range, an epidemiology that often involves food and animals, and a dramatically more severe and invasive presentation in immunocompromised adults, in particular in those with HIV. The prevalence of non-typhoidal Salmonella (NTS) bacteraemia has risen in many countries and is probably related to the increase in HIV infection. Although invasive disease caused by NTS has been recently reported from many African and Asian countries, the infection is relatively unknown in India.
So, in the current study the primer is chosen such that it detects other typhoidal as well as NTS. Analysis of the evolution of the genus Salmonella revealed that it is of monophyletic origin and that Salmonella serotypes last had a common ancestor with close relatives, such as Escherichia coli, some 100 to 150 million years ago [8]. During their separate evolution, Salmonella serotypes acquired many genes by phage or plasmid-mediated horizontal transfer. These genes now distinguish this genus from other bacteria and are thus obvious candidates for the development of DNA-based methods for identification of Salmonella serotypes. Two genetic regions on the S. enterica chromosome, Salmonella pathogenicity island 2 and the iroBC operon, indeed show this phylogenetic distribution [9].
The first genes of the iroA locus, designated iroBC, were identified in S. enterica serotype Typhi during a genetic screen for genes that are regulated by the iron response regulator Fur (Ferric Uptake Regulator). Regulation by Fur results in expression of iroBC under iron-limited growth conditions. In contrast, during growth under iron sufficiency expression is prevented by binding of the Fur-Fe21 repressor complex to a Fur DNA binding site in the iroB promoter region. However, the iroBC gene products show homology to proteins that have so far not been associated with iron uptake or defense against oxidative stress in other bacteria. IroB shows homology with bacterial glycosyl transferases, and IroC is a member of the ATP binding cassette (ABC) family of transport proteins[10].
Aim
To optimize PCR for iroB gene of S.enterica and evaluate its usefulness in the rapid diagnosis of typhoidal and non typhoidal salmonellosis.
Objectives
To optimize PCR for iroB gene of Salmonella enterica. and to compare iroB gene detection with that of fliC gene in diagnosis of typhoid fever.
Materials and Methods
The study was approved by Institute Ethics Committee, JIPMER, Pondicherry. This study was carried out between August 2009 and July 2011 on blood samples from 60 patients attending JIPMER Hospital, Pondicherry with clinical suspicion of enteric fever and non typhoidal salmonellosis. Two groups of 30 patients each were included in the study. Members of both sexes and all age groups were included. Group I consisted of blood samples from 30 patients whose blood culture grew Salmonella spp. Group II consisted of blood samples from 30 patients with clinical suspicion of Salmonellosis but blood cultures negative for Salmonella spp.
Patients with clinical suspicion of enteric fever who tested positive by Standard Agglutination test for Brucellosis, leptospirosis by IgM ELISA, rickettsial infections by Weil Felix test, malarial parasites by peripheral smear and QBC were excluded from the study. Three to five ml of blood were collected from patients coming to microbiology outpatient department in EDTA coated containers with informed consent. Pure culture of Salmonella Typhi H901 was used for standardisation of PCR.
DNA extraction: Extraction of DNA from whole blood was done as per procedure described by Haque et al., [11] with several modifications. Blood samples collected in EDTA containers inoculated into BHI broth and incubated for 6 h. One ml of supernatant was centrifuged at 10,000 rpm for 5 min. One ml of lysis buffer (0.2% Triton X100 in Tris HCl pH 8.0) was added to the pellet. The mixture was gently aspirated several times to effect hemolysis. The tube was centrifuged at 12,000 rpm for 6 min, the supernatant was discarded, and the procedure was repeated several times till a clear white pellet was observed. The pellet was washed twice with distilled water. After the removal of the supernatant, the pellet was resuspended in 20-30 μl of distilled water. The tubes were sealed, kept in boiling water for 20 min, and brought back to room temperature before being used as a template for PCR.
PCR Conditions: Primers targeting the flagellin gene of Salmonella Typhi were used after verifying against the database in the GenBank {GenBank accession no (AC): L21912} oligonucleotides ST3 (5’- ACTGCTAAAACCACTACT -3’) and ST4 (5’- TGGAGACTTCGGTTGCGTAG-3’) were used to amplify a 366 bp fragment corresponding to nucleotide 1060-1077 and 1407-1426 {(AC): L21912} [6]. Forward and reverse primers of iroB gene include oligonucleotides (5’-TGCGTATTCTGTTTGTCGGTCC-3’) and (5’TACGTTCCCACCATTCTTCCC-3’). With these primers, a 606-bp DNA fragment could be amplified from all S. enterica isolates [10]. A 25ml amplification mixture containing 10ml of master mix (Bangalore Genei, Bangalore) 7m of MilliQ water, 1ml fliC-F, 1ml fliC-R, 1ml iroB-F, 1ml iroB-R with 4ml of the extracted DNA were used. Primer details are in [Table/Fig-1]. The reaction was carried out in an Eppendorf Mastercycler (Eppendorf, Germany). The reaction mixture was submitted to touch down PCR of 25 cycles of 2 min denaturation at 94oC, annealing at 68oC for 1 min 15 sec and elongation at 72oC for 2 min followed by 20 cycles of initial denaturation of 94oC of 2 min, annealing at 55oC for 1 min and elongation at 72oC for 2 min. A final elongation of 10 min duration was done at 72oC.
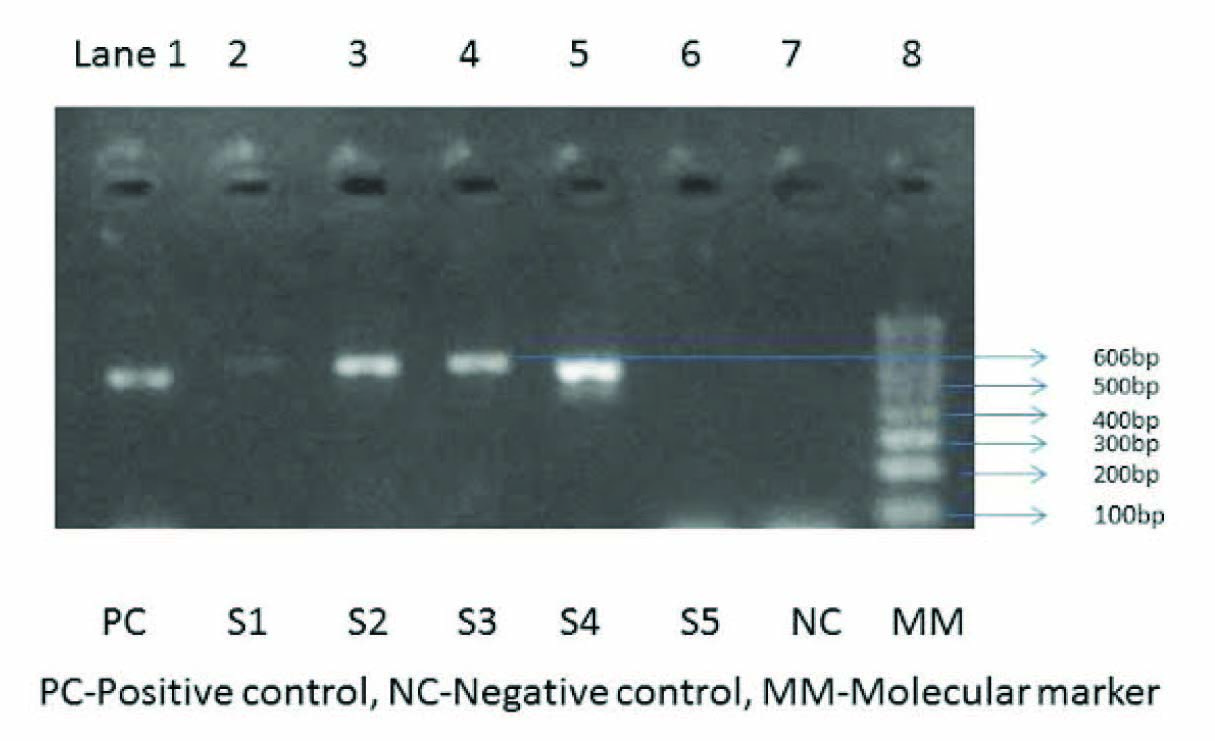
Detection of PCR products: A reaction mixture of 5ml was fractionated electrophoretically at 80V for 45 min in a 1.5% agarose gel containing 0.5mg of ethidium bromide. A positive control (Amplification mixture with DNA extracted from standard strain of Salmonella Typhi) and a negative control (Amplification mixture without DNA) were also included in each lot. The bands obtained were documented using a gel documentation system. With iroB primers, a 606-bp DNA fragment could be amplified from all S. enterica isolates. The fliC primers amplified a 366 bp fragment, in the flagellin gene of S.typhi.
Blood culture: Under aseptic precautions, 5-10 ml of blood from adults and 1-3ml of blood from paediatric age group was collected and inoculated into a biphasic medium consisting of Brain Heart Infusion (BHI) agar and BHI broth (Hi Media Laboratories Ltd, Mumbai). Bottles were checked daily for growth on the BHI slant or any turbidity in the BHI broth. If no visible growth occurred, blind subculture was performed. Any colony obtained on subculture was identified by biochemical tests as per standard recommendations and confirmed by specific antisera. (Denka Seiken Co. Ltd, Japan).
Results
In this study, the age of the patients ranged from 6 to 60 yrs with a mean of 26 yrs. Age distribution of the study population described in [Table/Fig-2]. Of the 60 patients included in the study, 30 grew Salmonella isolates in the blood culture. 24 isolates of Salmonella Typhi, 2 Salmonella spp, 3 Group B Salmonella and 1 Salmonella Paratyphi A. Of which 23 were positive for fliC gene by PCR. Over all 29 samples were positive for iroB gene [Table/Fig-3&Table/Fig-4] One sample which has grown S.typhi in blood culture and negative for fliC and iroB by PCR. This could be due to inhibitors in blood, mainly heme and leucocyte DNA, or added anticoagulants like EDTA. Two samples which were negative by blood culture were positive for fliC and iroB by PCR. PCR was also carried out on 10 blood samples which yielded Escherichia coli, Klebsiella pneumoniae, Enterobacter spp. in culture. All these samples were negative for fliC and iroB genes by PCR.
Discussion
Salmonellosis remains a major cause of morbidity and mortality in developing countries. Early diagnosis is not only necessary for detection of the cases but also to prevent fatal complications like intestinal perforation in case of enteric fever and focal infections like meningitis, osteomyelitis, septic arthritis and pneumonia in case of NTS. Age distribution of the population is needed to define the optimum age of immunization and choice of vaccines for public health programmes in developing countries. Highest attack rates occur in school going children and young adults. Invasive nontyphoidal salmonellosis shows a markedly contrasting age distribution showing a bimodal age distribution. In this study, though the number of cases of NTS is too small to extrapolate the pattern, they followed the same bimodal age distribution.
Generally isolation from blood culture is poor due to factors like prior antibiotic therapy, low bacterial count as low as 1CFU/ml [12]. Blood culture is usually negative in 30-65% of typhoid fever patients [13]. After standardization of PCR for flagellin gene of S.Typhi, it was evaluated against the results of blood culture in the absence of a better diagnostic test. PCR was able to detect flagellin gene in 25 out of the 60 patients (41.6%). The sensitivity of PCR was determined to be 96%. Several studies have reported high sensitivity of PCR of blood for S.Typhi [9,14]. PCR detected iroB gene in 31 out of the 60 blood samples (51.6%).The sensitivity of PCR was determined to be 96.6%.
However, initial research indicated that PCR has similar sensitivity to blood culture and lower specificity [15]. A prospective study of the concentration of S. Typhi in blood of typhoid fever patients showed a median value of 0.3 (range of 0.1 to 399) CFU/ml, well below current PCR-based detection limits [16]. Gordon et al [2] have shown that NTS in bacteremic patients are present at a similarly low concentration (1 CFU/ml).
To overcome the difficulties caused by the low numbers of Salmonella bacteria present in typhoid patient blood samples, pre-enrichment of bacteria is necessary prior to PCR detection. A recent study demonstrated that the 5 hour broth culture enrichment improved PCR sensitivity by 10 times for spiked blood, and 100 times for spiked stool samples [17]. In another study, ox bile tryptone soy broth was optimized for blood culture which caused lysis of cells to release the intracellular bacteria without inhibiting growth of S.Typhi, and then detected by PCR targeting fliC of S.Typhi [18]. In the current study, 6 hour BHI broth culture enrichment done prior to DNA extraction and by using iroB gene typhoidal and non typhoidal Salmonellae are detected which is advantageous over the more commonly targeted gene, fliC.
Primes used for PCR amplification of fliC and iroB genes
| Primers | Nucleotide sequence |
| fliC-F | 5’- ACTGCTAAAACCACTACT -3’ |
| fliC-R | 5’-TGGAGACTTCGGTTGCGTAG-3’ |
| iroB-F | 5’-TGCGTATTCTGTTTGTCGGTCC-3’ |
| iroB-R | 5’-TACGTTCCCACCATTCTTCCC-3’ |
Age details of the study population
| Age | Number(%) | Salmonella in blood culture |
| 1-10 | 5(8.3) | 4 |
| 11-20 | 20(33.3) | 13 |
| 21-30 | 16(26.6) | 8 |
| 31-40 | 11(18.3) | 2 |
| 41-50 | 2(3.3) | 1 |
| >50 | 6(10) | 2 |
Comparison of results between blood culture, fliC and iroB gene
| Blood culture | fliCgene positive | iroB gene positive |
| Salmonella Typhi | 24 | 23 | 23 |
| Salmonella Paratyphi A | 1 | 0 | 1 |
| Group B Salmonella | 3 | 0 | 3 |
| Salmonella spp | 2 | 0 | 2 |
Gel image after PCR for iroB gene of S.enterica
Conclusion
With iroB gene, additional cases of S.Paratyphi A and non typhoidal Salmonella were detected when compared to fliC gene. DNA extraction results were good after six hours of BHI enrichment of blood samples rather than directly from blood. In PCR positive and culture negative cases, antibiotic sensitivity results will not be known. Primers targeting drug resistance genes will serve the purpose.
[1]. MK Bhan, R Bahl, S Bhatnagar, Typhoid and paratyphoid feverLancet 2005 366(9487):749-62. [Google Scholar]
[2]. MA Gordon, AMK Kankwatira, G Mwafulirwa, AL Walsh, MJ Hopkins, CM Parry, Invasive Non-typhoid Salmonellae Establish Systemic Intracellular Infection in HIV-Infected Adults: An Emerging Disease PathogenesisClin Infect Dis 2010 50(7):953-62. [Google Scholar]
[3]. BS Reisner, GL Woods, Times to Detection of Bacteria and Yeasts in BACTEC 9240 Blood Culture BottlesJ Clin Microbiol 1999 37(6):2024-26. [Google Scholar]
[4]. G Durmaz, T Us, A Aydinli, A Kiremitci, N Kiraz, Y Akgun, Optimum Detection Times for Bacteria and Yeast Species with the BACTEC 9120 Aerobic Blood Culture System: Evaluation for a 5-Year Period in a Turkish University HospitalJ Clin Microbiol 2003 41(2):819-21. [Google Scholar]
[5]. J Wain, S Hosoglu, The laboratory diagnosis of enteric feverJ Infect Dev Ctries 2008 2(6):421-25. [Google Scholar]
[6]. S Khan, BN Harish, GA Menezes, NS Acharya, SC Parija, Early diagnosis of typhoid fever by nested PCR for flagellin gene of Salmonella enterica serotype TyphiIndian J Med Res 2012 136:850-54. [Google Scholar]
[7]. JA Crump, SP Luby, ED Mintz, The global burden of typhoid fever. BullWorld Health Organ 2004 82(5):346-53. [Google Scholar]
[8]. H Ochman, AC Wilson, Evolution in bacteria: Evidence for a universal substitution rate in cellular genomesJ Mol Evol 1987 26(1-2):74-86. [Google Scholar]
[9]. AJ Baumler, RM Tsolis, AW van der Velden, I Stojiljkovic, S Anic, F Heffron, Identification of a new iron regulated locus of Salmonella typhiGene 1996 18(1-2):207-13. [Google Scholar]
[10]. AJ Baumler, F Heffron, R Reissbrodt, Rapid detection of Salmonella enterica with primers specific for iroBJ Clin Microbiol 1997 35(1):1224-30. [Google Scholar]
[11]. A Haque, J Ahmed, JA Qureshi, Early detection of typhoid by polymerase chain reactionAnn Saudi Med 1999 19(4):337-40. [Google Scholar]
[12]. J Wain, TS Diep, VA Ho, AM Walsh, NTT Hoa, CM Parry, Quantitation of Bacteria in Blood of Typhoid Fever Patients and Relationship between Counts and Clinical Features, Transmissibility, and Antibiotic ResistanceJ Clin Microbiol 1998 36(6):1683-87. [Google Scholar]
[13]. R Chaudhry, BV Laxmi, N Nisar, K Ray, D Kumar, Standardisation of polymerase chain reaction for the detection of Salmonella typhi in typhoid feverJ Clin Pathol 1997 50(5):437-39. [Google Scholar]
[14]. MM Sánchez-Jiménez, N Cardona-Castro, Validation of a PCR for diagnosis of typhoid fever and salmonellosis by amplification of the hilA gene in clinical samples from Colombian patientsJ Med Microbiol 2004 53(1):875-78. [Google Scholar]
[15]. H Levy, S Diallo, SM Tennant, S Livio, SO Sow, M Tapia, PCR method to identify Salmonella enterica serovars Typhi, Paratyphi A, and Paratyphi B among Salmonella Isolates from the blood of patients with clinical enteric feverJ Clin Microbiol 2008 46(5):1861-66. [Google Scholar]
[16]. J Wain, VB Pham, V Ha, NM Nguyen, SD To, AL Walsh, Quantitation of bacteria in bone marrow from patients with typhoid fever: relationship between counts and clinical featuresJ Clin Microbiol 2001 39(4):1571-76. [Google Scholar]
[17]. ZA Bhutta, Current concepts in the diagnosis and treatment of typhoid feverBMJ 2006 333(7558):78-82. [Google Scholar]
[18]. L Zhou, AJ Pollard, A fast and highly sensitive blood culture PCR method for clinical detection of Salmonella enterica serovar TyphiAnn Clin Microbiol Antimicrob 2010 19(9):14 [Google Scholar]