Background: Iran has extended borders with high-TB burden countries (Afghanistan and Pakistan) and immigrations of these populations influences TB distribution in the region and threatens the control strategies. The aim of this study was to evaluate the extent of recent TB transmission among Iranian and Afghan cases.
Materials and Methods: Spoligotyping and 15-locus variable number tandem repeat (VNTR) typing were applied to genotype 102 MTB isolates (2009 to 2010). Phylogenetic relationships were analysed by two methods: a cluster-graph method and a minimum spanning tree (MST) method. Furthermore, evaluation of recent TB transmission was assessed with three indices including, RTIn, RTIn-1 and TMI.
Results: Using molecular typing, 35 different spoligotypes were detected among the studied isolates. Seventy seven cases (75.4%) were distributed into 10 clusters and the remaining 25 (24.5%) isolates had a unique pattern. The cluster sizes also ranged from 2 to 21 isolates. The most frequent spoligotype in our populations belong to Haarlem (n=30, 29.4%) followed by CAS (n= 29, 28.4%) and Beijing (n=16, 15.6%) lineages. The used indices give the following values: RTIn = 0.75, RTIn-1 = 0.65 and TMI = 0.24.
Conclusion: The low rate of TB transmission in our findings (24%) showed that the mode of TB transmission in Iran is mostly associated with reactivation of a previous TB infection and that recently a transmitted disease has a minor role. However, the increasing incidence of the intra-community transmission in recent years highlights the need for establishing new strategies for control of TB.
Background
Tuberculosis (TB) remains one of the major public health problems in Iran. According to the World Health Organization (WHO), the incidence rate of TB in Iran is approximately moderate, 21 cases per 100,000 populations [1]. However, the existence of extended borders with high-TB burden countries (Afghanistan and Pakistan) and immigrations of Afghans influences TB distribution in the region and threatens the control strategies.
Tracking the epidemiological events such as transmission dynamics of Mycobacterium tuberculosis (MTB) strains is one of the essential priorities for TB control strategy [2,3]. In this regard, several genotyping assays have been established, of which Spoligotyping and Mycobacterial Interspersed Repetitive Unit-Variable Number Tandem Repeat (MIRU-VNTR) were among the most frequently used [4-7]. Today, these methods, by detecting recent transmission and distinguishing between re-infections and relapses, have become a valuable tool for epidemiological study of TB. Although MIRU-VNTR is considered as a reliable and reproducible method, several studies shows that using the two techniques of MIRU-VNTR and Spoligotyping simultaneously could evaluate the genetic patterns of MTB strains in a much better manner[8] . Therefore in the current study, Spoligotyping and MIRU-VNTR assays was applied for genotyping and tracking the transmission dynamics of MTB strains isolated from different parts of Iran.
Materials and Methods
Clinical isolate and data collection
This cross-sectional study was conducted over a period of 1-year (from March 2009 to June 2010) in the Mycobacteriology Research Centre (MRC). MRC is the only national reference TB laboratory of Iran which is supervised by Swedish Institute for Infectious Disease Control. A total of 102 patients with culture-positive TB were included in the study. Demographical data of TB cases were collected using patient’s clinical records. The study was approved by the ethics committee of MRC.
Isolation of MTB
Samples from patient were decontaminated by Petroff’s method and were inoculated into Lowenstein-Jensen media [9]. Bacterial isolates were identified as MTB complex using spoligotyping and heat shock protein analysis [7,10].
DNA extraction
Genomic DNA of isolate was extracted as described by van Soolingen et al.,[11]. Briefly, 1.5 ml of the loopful culture was heated at 80°C for 20min to kill the bacteria. After centrifugation, the bacteria were re-suspended in TE buffer containing 1mg/ml of Lysozyme, and incubated for 1h at 37°C. Fifty mg proteinase K in 10% SDS was added to the mixture and incubated for 30min at 65°C. A further incubation with 80 μl 5M NaOH and CTAB/NaOH (CTAB in 0.7M NaOH) for 10min at 65°C was followed by adding an equal volume of chloroform-isoamyl alcohol (24:1, vol/vol) to the tube. Then, genomic DNA was precipitated with isopropanol at –20°C for 30min. After centrifugation, the pellet was washed with 70% ethanol and re-dissolved in TE buffer.
MIRU -VNTR typing
Standard 15-locus VNTR typing was applied to genotype the MTB clinical isolates as described by Supply et al.,[12] . The allelic diversity of each VNTR locus was evaluated by the Hunter – Gaston discriminatory index (HGDI), as described previously. The obtained MIRU patterns were compared with the MIRU-VNTR plus Database (http://www.miru-vntrplus.org) to determine MTB strain lineages and relatedness.
Spoligotyping
Spoligotyping was performed for all 102 MTB isolates according to the standard method [7] . Briefly, DR region of MTB strains was amplified using specific primers as described by Kemerbeek et al., [7]. The PCR amplicons were subsequently hybridized to a set of 43 different immobilized DR spacers covalently bound to a membrane. The hybridization signals were detected by chemiluminescence system after incubation with a streptavidin-peroxidase conjugate.
Analysis of VNTR and Spoligotyping allelic diversity and genetic relationships.
The allelic diversity and genetic relationships among the isolates of each VNTR locus and spoligopatterns were estimated using MIRU-VNTR plus software and SITVITWEB, respectively. Further confirmation of genotype classification was performed according to SITVITWEB. Cluster-graph and Minimum Spanning Tree (MST) was used to analyse the clonal pathogenic genotypes of studied strains [13]. Statistical analysis was carried out using MedCalc statistical software, version 11.6.0 (MedCalc Software bvba).
Estimation of transmission
Different approaches for estimation of transmission have been used in our analyses [14-16] . The first one was based on classical epidemiological assertions; 5 to 10% lifelong risk of developing TB following primary infection and the assumption that one smear-positive patient on average infects 13 persons per year. The second epidemiological assertion is based on recent transmission indices (RTI) i.e., RTIn = nc/n and RTIn-1 = (nc - c)/n, in which n is the total number of the studied cases, nc is the total number of cases in cluster (size two or greater) and c is the number of genotypes represented by at least two cases. Base on these indices, patients in cluster is considered as a recent transmission and non-cluster cases considered as a reactivation. These indices also have been referred to as the “n method” and the “n-1 method” respectively. The third approach is the transmission mutation index (TMI); TMI = μ (n-g+ν1)/ν1, where μ where is an independent estimate of the mutation rate of the genetic marker, and v1 is the number of single-step mutation events inferred from the data. These indices are variable between 0 (the least recent transmission) to 1 (maximum genotypes transmission).
Results
According to the demographic characteristics of studied populations, 73 (71.5%) of investigated cases were Iranian and the remaining 29 (28.4%) cases were immigrants [Table/Fig-1]. Patients in the group aged > 60yr had the highest number of cases, followed by those aged 15-30yr. In the terms of gender, 51.7% of immigrants were female and 48.2% were males; while in Iranian patients 32.8% were females and 67.1% were males [Table/Fig-1].
As shown in [Table/Fig-2], all clinical isolates of MTB were successfully fingerprinted by 15-locus MIRU-VNTR and spoligotyping. Using spoligotyping, 35 different spoligotypes and 3 orphan patterns (4.10%) were detected among the studied isolates. Seventy seven cases (75.4%) were distributed into 10 clusters and the remaining 25 (24.5%) isolates had a unique pattern. Five clusters belonged to the Iranian cases only. Furthermore, of the 78 clustered isolates, 19 (24.35%) belonged to the immigrants and 59 (75.64%) were Iranians. The cluster sizes also ranged from 2 to 21 isolates. Additionally, the most frequent spoligotype in our populations belong to Haarlem (n=30, 29.4%) followed by CAS (n= 29, 28.4%) and Beijing (n=16, 15.6%) lineages [Table/Fig-2].
By 15-loci MIRU-VNTR genotyping, closely related patterns were observed between MTB strains. Moreover, a comparison of the MIRU patterns obtained with the international MIRU-VNTR plus database (http://www.miru-vntrplus.org) showed that none of the strains out of the 186 present in this database matched our patterns. The discriminatory power of MIRU-VNTR typing for all isolates was approximately high (HGDI=0.60). Based on the discriminatory index, all MIRU loci were designated as highly discriminative ( h > 0.6) except MIRU04.
Cluster-graph analysis and MST was performed by Spoltools and MIRU-VNTRplus sites, respectively.
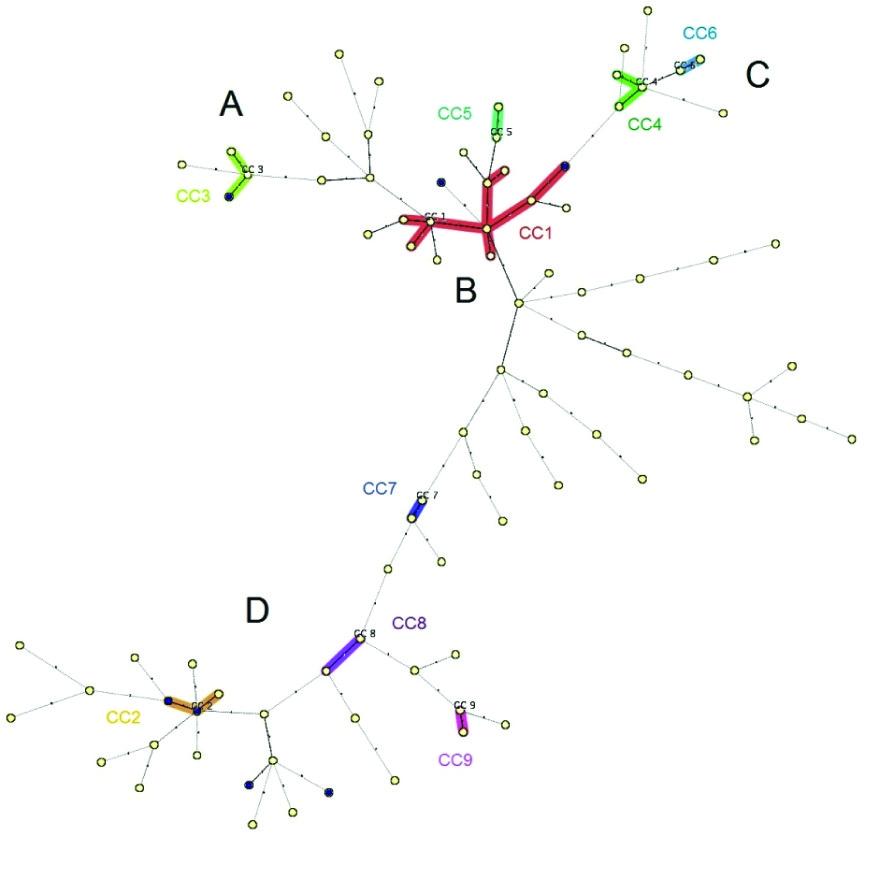
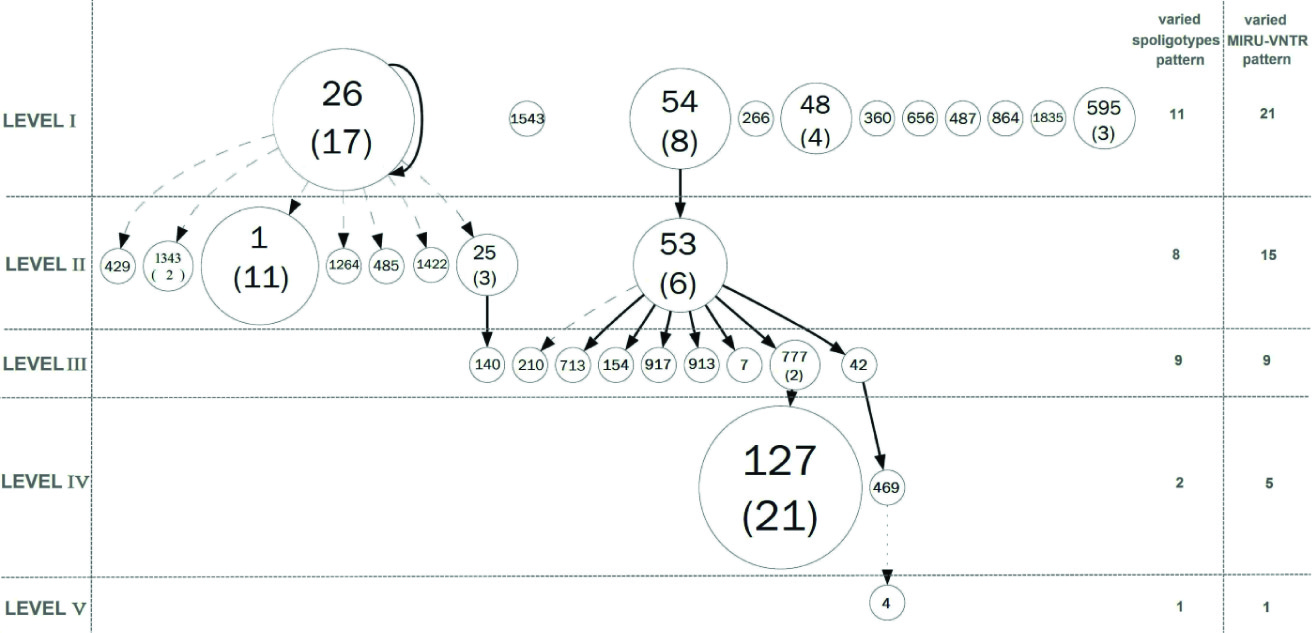
As shown in [Table/Fig-3] , combined numerical analysis of spoligotyping and VNTR data underlined four well-defined branches (A, B, C and D), rising from a central cluster called B. Branch A mostly contains genotypes from Beijing families; branch B shows genotypes from Haarlem and MANU families; branch C contains genotypes from T, and branch D includes CAS genotypes. [Table/Fig-4] also illustrates the complete cluster-graph, with all different spoligotypes that occurred in our study. Each node represents distinct genotype, and the size of nodes is proportional to the corresponding cluster size. Moreover, the levels (I to V) indicate the sense of evolution through mutation events. It is supposed that isolates in the cluster-graph belonged to the same epidemic.
Using the indices previously described, data set gives the following values: RTIn = 0.75, RTIn-1 = 0.65 and TMI = 0.24, with n = 102 isolates, nc = 77 isolates in cluster and c = 10 clusters. The number v1 is equal to the number of genotypes g in the data set minus the number of connected components, which is calculated using the cluster-graph.
Discussion
Based on the recent WHO report, the incidence rate of TB in Iran was approximately moderate, 21 cases per 100,000 populations [1] . However, the existence of extended borders with countries where TB is endemic (Afghanistan and Pakistan) and immigrations of these populations influenced TB distribution in the region and threatens the control strategies.
In the current study, the major identified isolates of MTB were characterised with Haarlem (n=30, 29.4%) and CAS (n=26, 25.4%) lineages [Table/Fig-2]. The Haarlem family was first identified by Kremer et al., in Netherlands, but soon after they have been described by many countries, i.e. Iran, Pakistan, Turkey, Iraq [17-20] . In addition, it was indicated that this particular genotype of MTB has an epidemiological potential to be transmitted and disseminated throughout the world. Farnia et al., has demonstrated a linkage patterns between 22 Iranian and Afghan patients with a Haarlem spoligopattern in which they were grouped into the same clusters [17] . They showed that patients in these clusters were either living in the same region or had intra-family relationship. However, in our study, the direct transmission link could not be established due to inadequate demographic data.
Another identified family of MTB strains in our populations was the CAS lineage (n: 29; 28.4%). In the previous reports, this lineage has been shown as a predominant strain in the central Asian countries where TB is endemic, i.e. Afghanistan and Pakistan [21] . Obviously, Iran is neighboring to these countries and shares long geographical borders (1500 km) with them. Consequently, frequent movement of ethnic groups may be one of the factors to explain the prevalence of the CAS family in Iran.
In other hand, according to the molecular typing results, about 75.4% of our isolates were distributed into 10 clusters with different range of sizes (from 2 to 21 isolates, [Table/Fig-2]). Although, this proportion of clustered strains was in the range with those reported by neighboring countries i.e. Turkey (79%) and Pakistan (63%), but the low mean of cluster size in spite of the high level of clustering seems to show a low level of recent transmission[19,21] . Therefore, the possibilities of reactivation of a previous infection with particular MTB strains were highlighted.
Moreover, based on the transmission indices, the result of TMI shows a recent transmission rate of 24%, instead of 75% and 65% of recent transmission estimated with RTIn and RTIn-1, respectively. Although, the recent transmission rate calculated with these methods demonstrate a considerable difference, comparing RTI and TMI indices shows a lower ongoing transmission rate with TMI method (24%). These indices are based on the same suppositions about the potential epidemiological link between patients sharing the same MTB genotypes. But as described by earlier studies, the major limitation with RTI techniques is their failure to account for strains diversity and mutation [13] . In addition, in the previous studies conducted in Iran, the rate of intra-community transmission was calculated to be 41% based on the combined spoligotyping and RFLP genotyping [22]. The difference between our observations and the previous study may be due to different approaches. In this regard, several investigations show that using the two methods of spoligotyping and MIRU-VNTR typing concurrently could identify outbreak episodes and rate of ongoing transmission in a much better manner [23].
However, some limitations of this study should be considered for interpretation. First, it cannot represent the accurate mode of the TB transmission in the Iran because the magnitude of disease is not yet known in many areas of the country. Second, the potential influence patient’s characterisations could not be analysed due to the limited information obtained from the clinical record. Third, the number of isolates used for this analyses was relatively small.
Demographic characterizations of Iranian and Afghan cases
| Characteristic | Afghan (%) (n = 29) | Iranian (%) (n = 73) |
| Gender |
| Female | 15 (51.72) | 24 (32.87) |
| Male | 29 (48.27) | 73 (67.12) |
| Age group |
| <15 | 0 (0) | 3 (4.10) |
| 15-30 | 18 (62.06) | 5 (6.84) |
| 31-45 | 8 (27.58) | 13 (17.80) |
| 45-60 | 2 (6.89) | 19 (26.02) |
The genotype patterns of MTB strains among Iranian and Afghan patients.
| Genotypes | SIT | Total (% of all) | Afghans (n=29) | Iranian (n=73) | MIRU-VN TR pattern |
| H | 127 | 21 | 7(24.13) | 14(19.17) | 4 |
| 777 | 2 | 0 | 2(2.73) | 1 |
| CAS1_DELHI | 26 | 17 | 6(20.68) | 11(15.06) | 4 |
| 25 | 3 | 0 | 3(4.10) | 1 |
| 1343 | 2 | 1(3.44) | 1(1.36) | 2 |
| Beijing | 1 | 11 | 5(17.24) | 6(8.21) | 3 |
| MANU2 | 54 | 8 | 0 | 8(10.95) | 7 |
| T1 | 53 | 6 | 0 | 6(8.21) | 6 |
| H3 | 48 | 1 | 1(3.44) | 3(4.10) | 2 |
| BOV | 595 | 3 | 0 | 3(4.10) | Undet-ected |
| Total clusters | | 77(75.49) | 20(68.96) | 57(78.08) | |
| Unique Iranian strains | 140-1543-794-210-154-917-913-7-656-487-4-485-392 | 13 | - | 13(17.80 | |
| Unique Afghans strain | 429-713-266-360-469-42-1264-1422-864 | 9 | 9(31.03) | - | |
| Orphan | unknown | 3 | 0 | 3(4.10) | |
| Total | | 25(24.50) | 9(31.03) | 16(21.91) | |
Minimum Spanning Tree. Combined numerical analysis of spoligotyping and VNTR data underlined four well-defined branches (A, B, C and D), rising from a central cluster called B.
Cluster-graph drawn from spoligotyping data. Each node represents distinct genotype, and the size of nodes is proportional to the corresponding cluster size.
Conclusion
In conclusion, the low rate of TB transmission in our findings (24%) showed that the mode of TB transmission in Iran is mostly associated with reactivation of a previous TB infection and that recently a transmitted disease has a minor role. However, the increasing incidence of the intra-community transmission in recent years highlights the need for establishing new strategies for control of TB. Our results also confirmed that the use of cluster-graphs from spoligotyping and MIRU-VNTR typing data to derive a TMI provides more sensitive results to identify the severity of outbreak episodes.
[1]. World Health Organization (WHO), Global Health Observatory Data Repository.Seehttp://apps.who.int/ghodata 2011. [Google Scholar]
[2]. PF Barnes, MD Cave, Molecular epidemiology of tuberculosis.New England J of Med. 2003 349(12):1149-56. [Google Scholar]
[3]. D Van Soolingen, K Kremer, E Vynycky, New perspectives in the molecular epidemiology of tuberculosis.Mycobacteria and TB Issues Infect Dis. 2004 (2):17-45. [Google Scholar]
[4]. J Van Embden, MD Cave, JT Crawford, J Dale, K Eisenach, B Gicquel, Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology.J of clini microbiol. 1993 31(2):406-09. [Google Scholar]
[5]. LS Cowan, L Mosher, L Diem, JP Massey, JT Crawford, Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J of clini microbiol. 2002 40(5):1592-1602. [Google Scholar]
[6]. P Supply, E Mazars, S Lesjean, V Vincent, B Gicquel, C Locht, Variable human minisatellitelike regions in the Mycobacterium tuberculosis genome. Moleul microbiol. 2000 36(3):762-71. [Google Scholar]
[7]. J Kamerbeek, L Schouls, A Kolk, M Van Agterveld, D Van Soolingen, S Kuijper, Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J of clin microbiol. 1997 35(4):907-14. [Google Scholar]
[8]. C Sola, I Filliol, E Legrand, S Lesjean, C Locht, P Supply, Genotyping of the Mycobacterium tuberculosis complex using MIRUs: association with VNTR and spoligotyping for molecular epidemiology and evolutionary genetics.Infec, Genetics and Evol. 2003 3(2):125-33. [Google Scholar]
[9]. S Petroff, A new and rapid method for the isolation and cultivation of tubercle bacilli directly from the sputum and feces. The J of experim med. 1915 21(1):38-42. [Google Scholar]
[10]. L Dvorska, M Bartos, G Martin, W Erler, I Pavlik, Strategies for differentiation, identification and typing of medically important species of mycobacteria by molecular methods.Vet Med Praha. 2001 46:309-28. [Google Scholar]
[11]. D van Soolingen, PE de Haas, PW Hermans, JD Van Embden, DNA Fingerprinting of Mycobacterium tuberculosis.Methods Enzymol. 1994 235:196-205. [Google Scholar]
[12]. P Supply, C Allix, S Lesjean, M Cardoso-Oelemann, S Rüsch-Gerdes, E Willery, Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis.J of Clin Microbiol. 2006 44(12):4498-510. [Google Scholar]
[13]. V Guernier, C Sola, K Brudey, J-F Guégan, N Rastogi, Use of cluster-graphs from spoligotyping data to study genotype similarities and a comparison of three indices to quantify recent tuberculosis transmission among culture positive cases in French Guiana during a eight year period. BMC Infect Dis. 2008 (8):46 [Google Scholar]
[14]. MM Tanaka, AR Francis, Methods of quantifying and visualising outbreaks of tuberculosis using genotypic information.Infect, Genetics and Evol. 2005 5(1):35-43. [Google Scholar]
[15]. J Glynn, E Vyonycky, P Fine, Influence of sampling on estimates of clustering and recent transmission of Mycobacterium tuberculosis derived from DNA fingerprinting techniques.Amer J of Epidemiol. 2002 149(4):366-71. [Google Scholar]
[16]. M Murray, Sampling bias in the molecular epidemiology of tuberculosis.Emerg Infect Dis. 2002 8:363 [Google Scholar]
[17]. P Farnia, MR Masjedi, M Mirsaeidi, F Mohammadi, Ghanavi Jallaledin, V Vincent, Prevalence of Haarlem I and Beijing types of Mycobacterium tuberculosis strains in Iranian and Afghan MDR-TB patients.J of Infect. 2006 53(5):331-36. [Google Scholar]
[18]. M Tanveer, Z Hasan, AR Siddiqui, A Ali, A Kanji, S Ghebremicheal, Genotyping and drug resistance patterns of M. tuberculosis strains in Pakistan.BMC Infect Dis. 2008 8:171 [Google Scholar]
[19]. O Kisa, G Tarhan, S Gunal, A Albay, R Durmaz, Z Saribas, Distribution of spoligotyping defined genotypic lineages among drug-resistant mycobacterium tuberculosis complex clinical isolates in ankara, turkey. PloS One. 2012 7(1):e30331 [Google Scholar]
[20]. MA Merza, AM Salih, First insight into the genetic diversity of Mycobacterium tuberculosis strains from patients in Duhok, Iraq.Internat J of Mycobac. 2012 1:13-20. [Google Scholar]
[21]. Z Hasan, M Tanveer, A Kanji, Q Hasan, S Ghebremichael, R Hasan, Spoligotyping of Mycobacterium tuberculosis isolates from Pakistan reveals predominance of Central Asian Strain 1 and Beijing isolates. J Clin Microbiol. 2006 44(5):1763-8. [Google Scholar]
[22]. MR Masjedi, M Varahram, M Mirsaeidi, M Ahmadi, M Khazampour, P Tabarsi, The Recent-Transmission of Mycobacterium tuberculosis Strains among Iranian and Afghan Relapse Cases: a DNA-fingerprinting using RFLP and spoligotyping.BMC Infect Dis. 2008 8:109 [Google Scholar]
[23]. W Lu, B Lu, Q Liu, H Dong, Y Shao, Y Jiang, Genotypes of Mycobacterium tuberculosis isolates in rural China: using MIRU-VNTR and spoligotyping methods.Scand J Infect Dis. 2014 (46)(2):98-106. [Google Scholar]