MRSA strains in the community were earlier distinguished from those in a healthcare setting by the presence of pvl genes (genes coding for Panton-Valentine leukocidin) in the former, and the display of multi-drug resistance by the latter [4]. However, several recent studies show that strains isolated from healthcare institutions possess pvl genes, and strains isolated from the community exhibit multi-drug resistance [5–9]. Some studies also show that strains isolated from the community are more virulent than those isolated from healthcare setups [10,11].
At present, sustained research that examines the influence of exposure to hospital environment on MRSA carriage is lacking in India, and, to the best of our knowledge, no Indian study has yet shown if there is any significant difference in the MRSA carrier rates, antibiotic resistance and pathogenicity between those exposed to a hospital environment (like nurses) and those in the community. This cross-sectional study was carried out to examine the influence of exposure to hospital environment on MRSA carriage by comparing the prevalence of MRSA carriage among healthy individuals exposed to a hospital environment and those not exposed. The antimicrobial resistance patterns of MRSA isolates in the two groups, and the presence of genes encoding five extracellular pathogenicity determinants (pvl, sea, seb, tsst 1 and hly a) are also compared.
A previous study done on the same samples by the same research team evaluated the prevalence of inducible clindamycin resistance in S. aureus isolated from carriers exposed and not exposed to hospital environment [12].
Materials and Methods
Study Duration and Participants
This study was carried out in the town of Tumkur in southern India. It was conducted between September 2010 and February 2012, with sample collection and initial phenotypic testing done between September 2010 and May 2011, and molecular analysis done between December 2011 and February 2012.
The group exposed to hospital environment comprised 119 nursing students from our hospital aged between 18-23 years, who attended daily eight hour rotations in various hospital departments. Samples were collected only from third and fourth year nursing students as they had daily postings in clinical departments. These students had various duties, including monitoring patients, administering drugs, carrying out various procedures (like inserting and removing intravenous cannulas and catheters), and updating patient and ward records.
The group not exposed to a hospital environment comprised 100 age matched pharmacy students from Siddaganga Pharmacy College, Tumkur. None of the students in this group had a history of hospitalisation or regular visits to a hospital in the last six months. Students from this pharmacy college were selected based on support for this study from the principal and the students.
None of the participants in either group had a history of illness or treatment with an antibiotic in the last six months.
Only students were included in our study (and made representative of the groups exposed and not exposed to hospital environment) because we wished to educate them about MRSA carriage, inform them of their carrier status, and impress upon them the need to take appropriate precautions to prevent transmission of MRSA.
Informed written consent was obtained from all participants. Each participant completed a standardised questionnaire that included the participant’s age, gender, and medical history over the preceding six months, including hospitalisation and antibiotic intake.
This study was approved by the Institutional Ethical Committee.
Bacterial Isolates
Swabs were collected from the nasal vestibules, throats, palms and web-spaces of the participants, and S. aureus identified phenotypically by growth on 5% sheep blood agar, 10% mannitol salt agar (HiMedia, Mumbai), Gram’s stain, catalase test, and slide and tube coagulase tests [13–15].
Isolates resistant to methicillin were identified phenotypically by growth on oxacillin screen agar [16], and by the cefoxitin disc diffusion test [16,17]. Quality control strains, MRSA ATCC 43300 and methicillin-sensitive S. aureus (MSSA) ATCC 25923, were used as positive and negative controls respectively. The isolates were inoculated into semisolid agar medium.
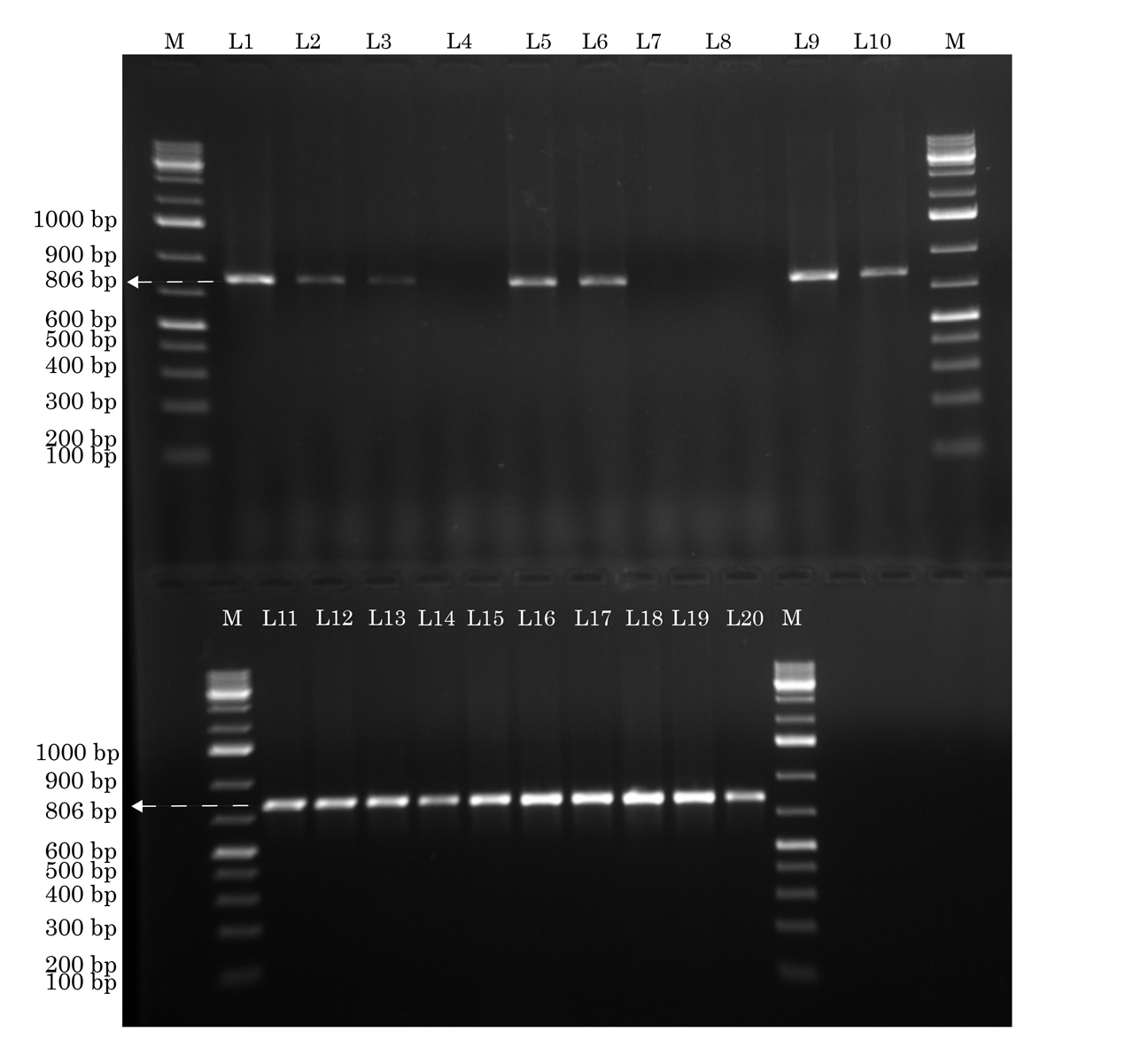
Resistance to methicillin was verified by detection of the mecA gene [Table/Fig -1] by conventional PCR performed on an Eppendorf Pro Gradient thermal cycler (Eppendorf AG, Germany) [18]. The primer pair was designed using primer BLAST and Gene Runner software version 3.05 (Hastings Software Inc., USA). The quality control strain used was ATCC 700699. In the case of multiple isolates acquired from the same participant, the first detected isolate of MRSA was included in the study.
Identification of mecA gene by conventional PCR. The lanes marked M to the left and right contain molecular weight markers. Lanes L1-L3, L5, L6, L9-L20 show the mecA gene (806 base pairs), while lanes L4, L7, L8 do not Forward primer: tccaggaatgcagaaagac. Reverse primer: ctggtgaagttgtaatctgga. Annealing temperature: 58°C
Antibiotic Resistance Testing
Antibiotic resistance of the isolates was assessed using standard disc diffusion tests as per CLSI guidelines [17]. Antibiotic discs (HiMedia, Mumbai) of penicillin (10 μg), cotrimoxazole (trimethoprim – 1.25 μg; sulfamethoxazole – 23.75 μg), ciprofloxacin (5 μg), gentamicin (10 μg), netilmicin (30 μg), tetracycline (10 μg), erythromycin (15 μg), clindamycin (2 μg), linezolid (30 μg), vancomycin (30 μg) and teicoplanin (30 μg) were used.
Presence of Extracellular Pathogenicity Determinants
The MRSA isolates were examined for the presence of five extracellular pathogenicity determinants: lukS of pvl [5], sea, seb, tsst 1 and hly a [Table/Fig 2,3,4,5and6]. Conventional PCR was performed for each gene [18]. PCR was performed on an Eppendorf Pro Gradient thermal cycler (Eppendorf AG, Germany).The primer pairs were designed using primer BLAST and Gene Runner software version 3.05 (Hastings Software Inc., USA). Primer pairs were highly specific in BLAST analysis with very little specificity to unintended targets. The PCR products were run on 1.5 % agarose gel stained with ethidium bromide to visualise the products under an ultraviolet transilluminator. Lanes with bright orange bands in the required regions were taken as positives for that particular gene. The gels were photographed in the gel documentation system. Quality control strains for PCR detection of pathogenicity factors were ATCC 49775 for lukS of pvl, ATCC 29213 for sea, FRI 722 for seb, TSST Jap for tsst 1, and ATCC 6538p for hly a.
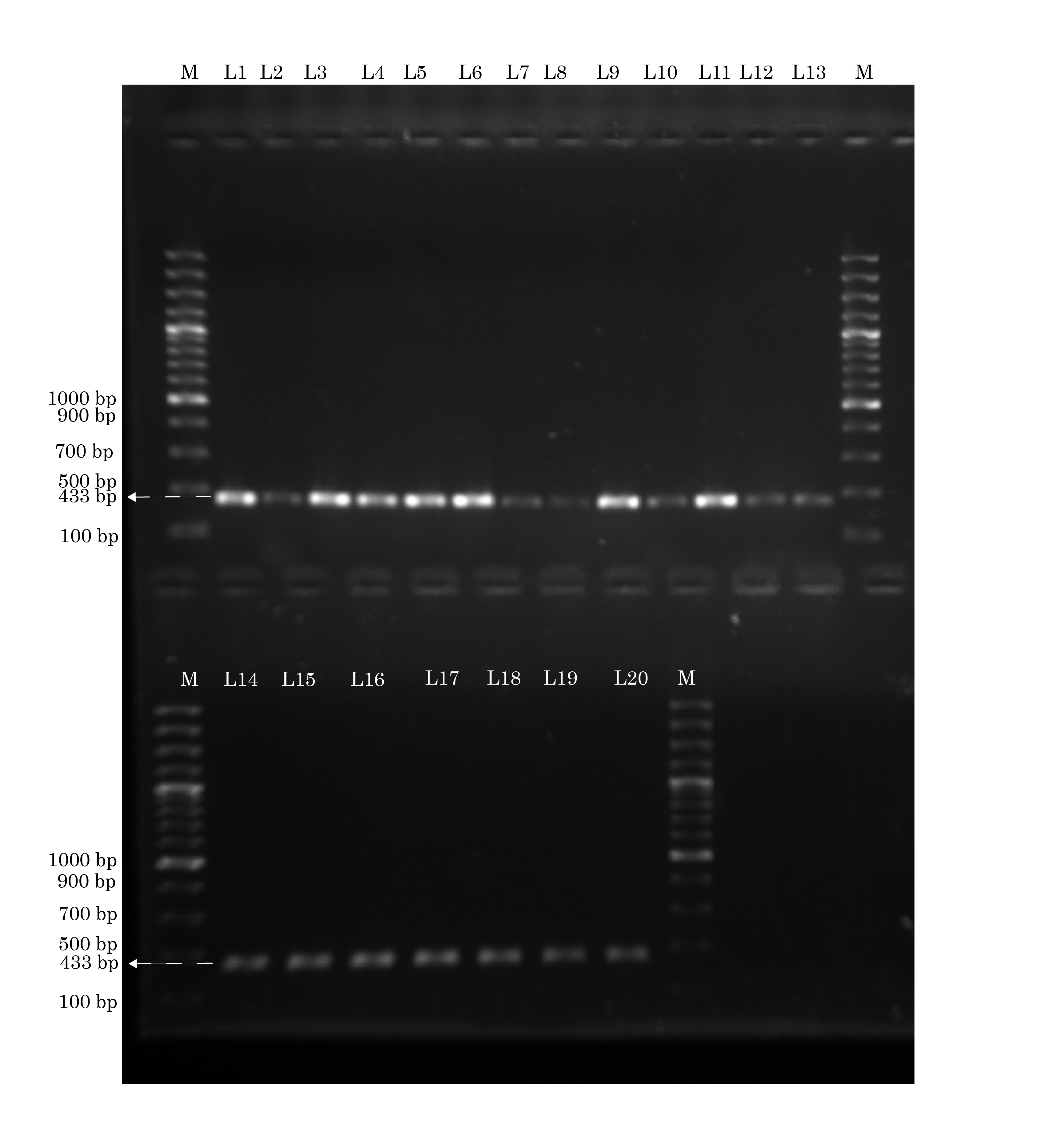
Identification of lukS of pvl by conventional PCR. The lanes marked M to the left and right contain molecular weight markers. All lanes L1-L20 show lukS of pvl (433 base pairs) Forward primer: gcagctcaacatatcacacc. Reverse primer: tgtttccagcagctttgag. Annealing temperature: 57°C
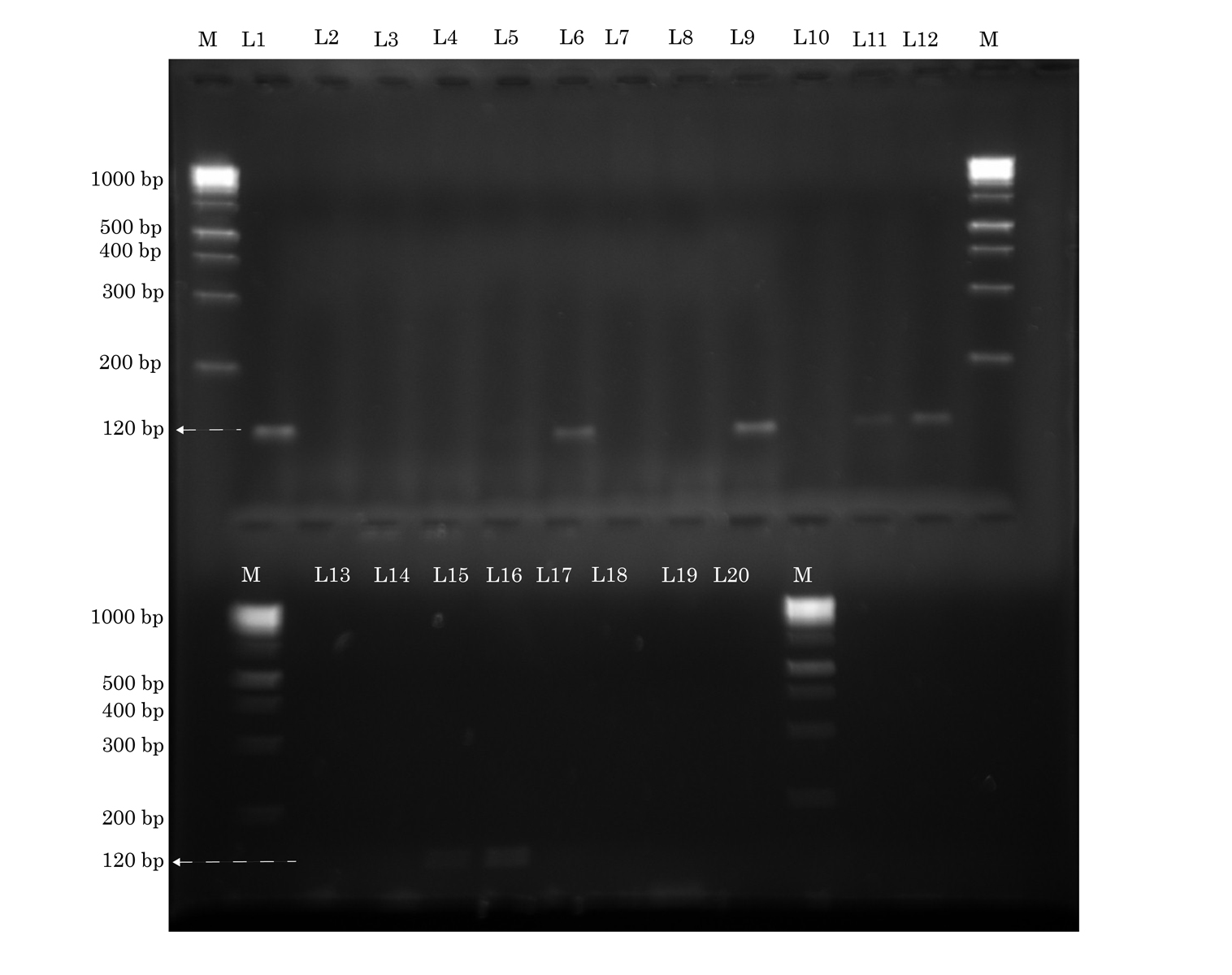
Identification of sea gene by conventional PCR. The lanes marked M to the left and right contain molecular weight markers. Lanes L1, L6, L9, L11, L12, L15, L16 show sea (120 base pairs) Forward primer: gttatcaatgtgcgggtg. Reverse primer: ggcactttttctattcgg. Annealing temperature: 58°C
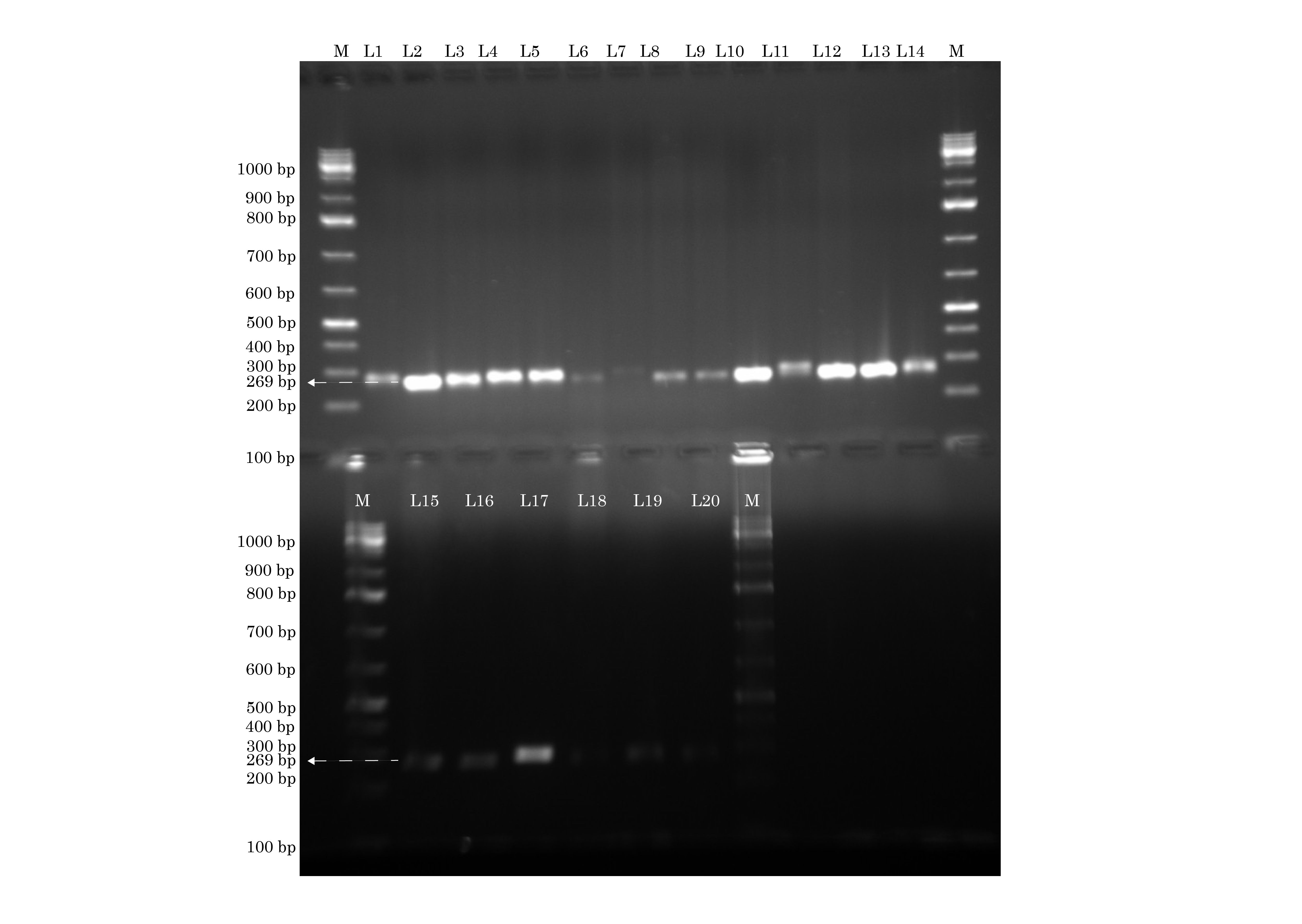
Identification of seb gene by conventional PCR. The lanes marked M to the left and right contain molecular weight markers. All except lanes L6, L7, L18 show seb (269 base pairs) Forward primer: tgtatggtggttaactgagc. Reverse primer: tgcaggcatcatgtcatac. Annealing temperature: 59°C
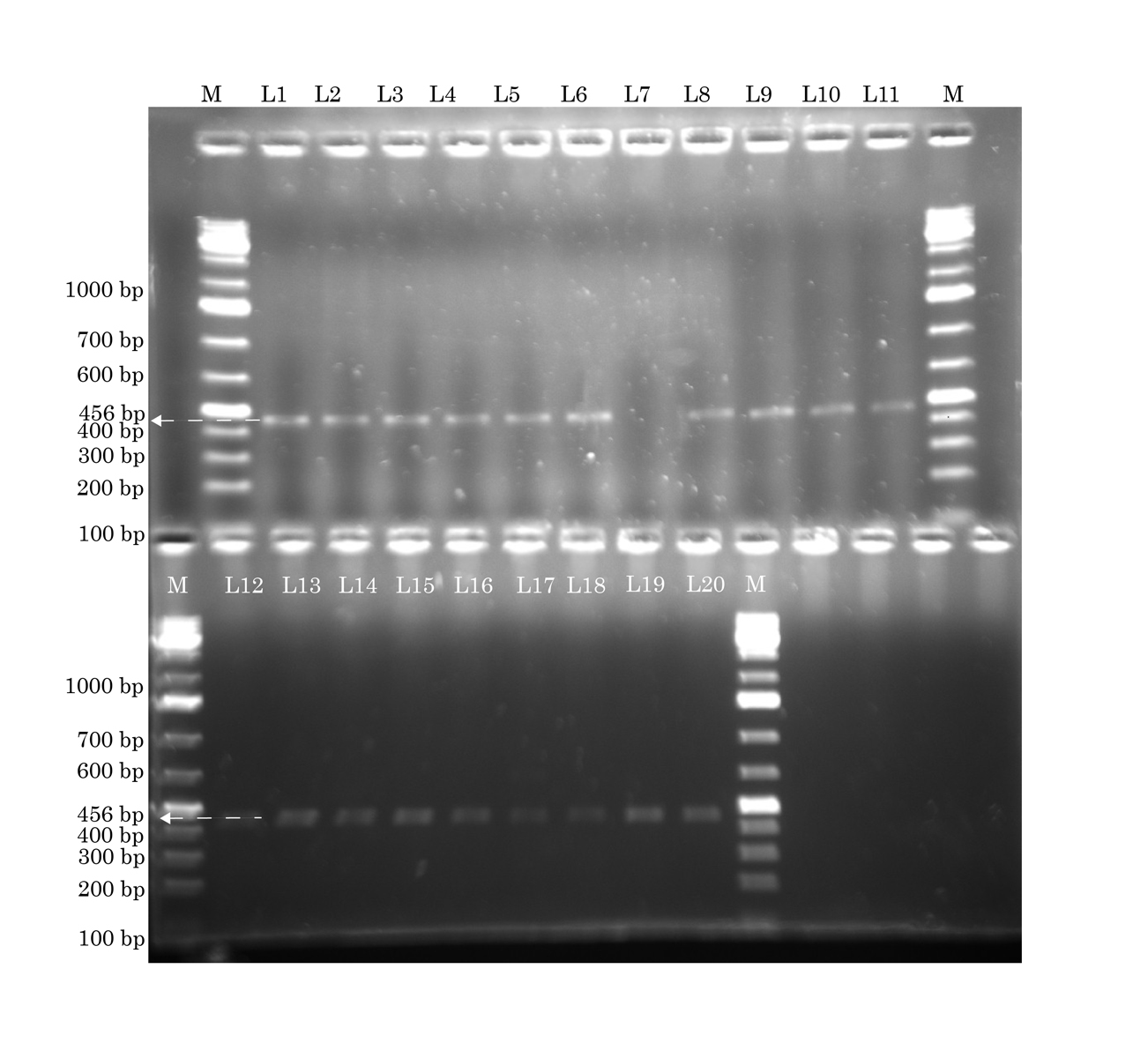
Identification of tsst 1 gene by conventional PCR. The lanes marked M to the left and right contain molecular weight markers. All except lane L7 show tsst 1 (456 base pairs) Forward primer: ccctgttcccttatcatct. Reverse primer: tccatgtatttgagttactg. Annealing temperature: 59°C.
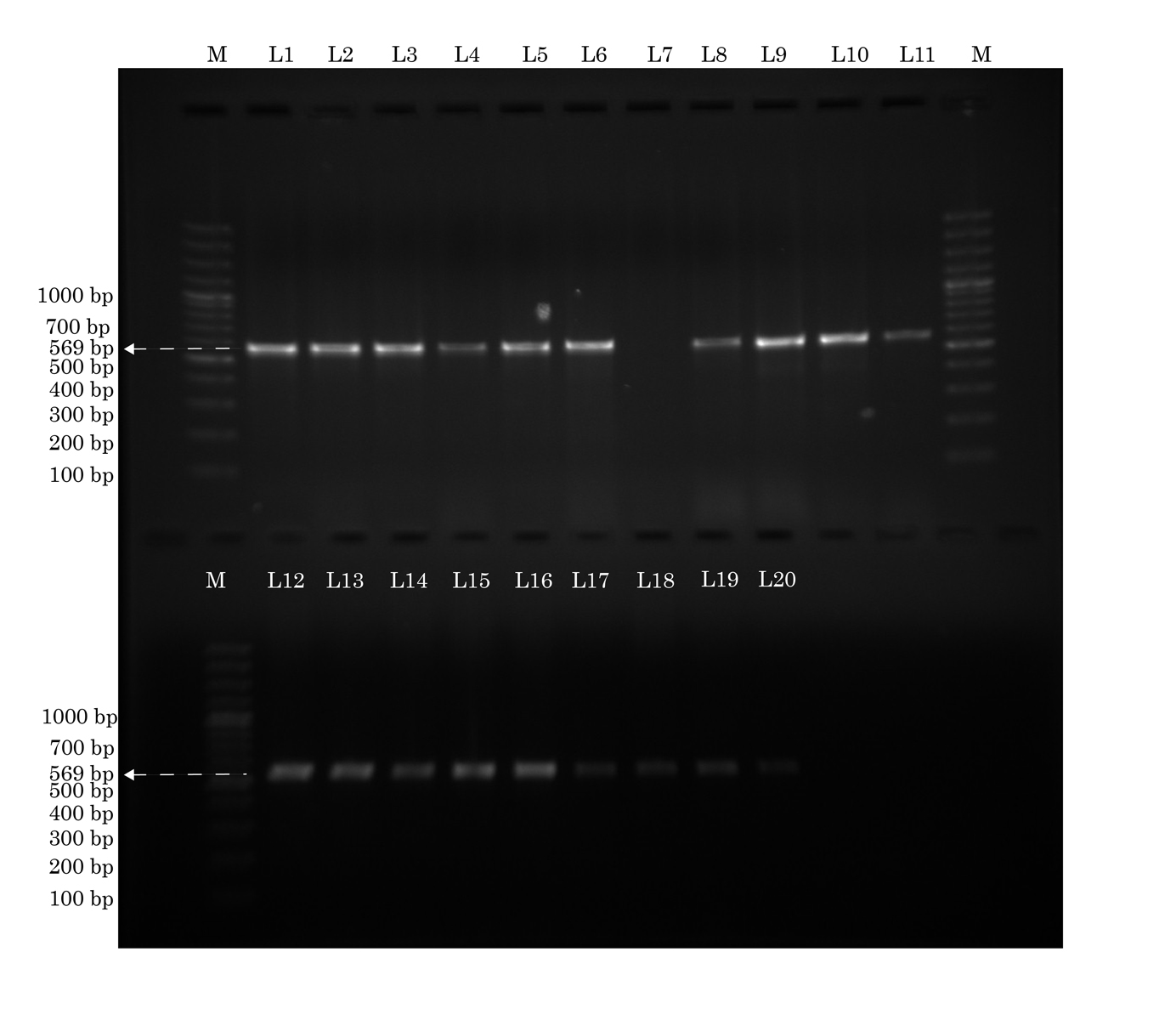
Identification of hly a gene by conventional PCR. The lanes marked M to the left and right contain molecular weight markers. All except lane L7 show hly a (569 base pairs) Forward primer: caattggtagtcatcacgaac. Reverse primer: gcgaagaaggtgctaaca. Annealing temperature: 57°C
Statistical Analysis
Statistical analysis of the association of exposure to hospital environment with MRSA colonization, as well as the association between MRSA isolated from hospital exposed participants and carriage of sea and seb genes, was carried out using Fisher’s exact test. Two-tailed p-values were calculated. All statistical analyses were done using GraphPad InStat version 3.10 for Windows 7 (GraphPad Software, USA).
Results
A total of 36 (30.3%) students were found to be carriers of S. aureus in the group exposed to hospital environment, and 34 (34%) in the group not exposed.
Of the 36 S. aureus isolates in the exposed group, 11 (30.6%) carried the mecA gene and were resistant to methicillin. In the non-exposed group, 4 (11.8%) of the 34 isolates carried the mecA gene. Therefore, of 119 participants studied in the exposed group, 11 carried MRSA (9.2% carrier rate), and of the 100 participants studied in the non-exposed group, four carried MRSA (4% carrier rate). The association of exposure to hospital environment with MRSA carriage was not statistically significant (p = 0.1795; p > 0.05).
The antibiotic resistance patterns of MRSA isolates from the exposed and non-exposed groups are given in [Table/Fig-7].
Antibiotic resistance patterns of MRSA isolated from the exposed and non-exposed groups
| Antibiotic | Exposed group | Non-exposed |
|---|
| Number (%) | Number (%) |
|---|
| Penicillin | 11 (100%) | 4 (100%) |
| Vancomycin | 0 (0%) | 0 (0%) |
| Teicoplanin | 0 (0%) | 0 (0%) |
| Linezolid | 0 (0%) | 0 (0%) |
| Erythromycin* | 7 (63.6%) | 1 (25%) |
| Clindamycin† | 3 (27.3%) | 1 (25%) |
| Cotrimoxazole | 7 (63.6%) | 1 (25%) |
| Netilmicin | 0 (0%) | 0 (0%) |
| Gentamicin* | 3 (27.3%) | 1 (25%) |
| Ciprofloxacin* | 7 (63.6%) | 3 (75%) |
| Tetracycline | 0 (0%) | 0 (0%) |
* Isolates with intermediate resistance were included with the resistant ones.
† Resistance to clindamycin was induced by erythromycin. These isolates showed a positive D-test.
The number of MRSA isolates in each group carrying various pathogenicity factors is given in [Table/Fig-8].
Number of MRSA isolates in each group carrying various pathogenicity factors
| Exotoxin | Exposed group | Non-exposed |
|---|
| Number (%) | Number (%) |
|---|
| LukS | 11 (100%) | 4 (100%) |
| sea | 7 (63.6%) | 0 (0%) |
| seb | 10 (90.9%) | 3 (75%) |
| tsst 1 | 11 (100%) | 4 (100%) |
| hly a | 11 (100%) | 4 (100%) |
Discussion
Staphylococcus aureus carrier rates in the two study groups were similar. Staphylococcus aureus carrier rates in the two study groups are similar. This was expected as S. aureus is a part of normal human microflora [2]. Also, the S. aureus carrier rates we obtained in our study fall within the normal range of 30% - 50% [2].
The percentages of S. aureus isolates that were methicillin resistant in the two study groups (30.6% in the exposed group and 11.8% in the non-exposed group) are slightly less than those obtained in a previous comparative study in Jordan, in which 38% and 18% of S. aureus were methicillin resistant in the exposed and non-exposed groups respectively (also comprising students) [19]. The MRSA carrier rate in the exposed group (9.2%) is more than that obtained in two previous Indian studies on healthcare workers (6.6% and 2.5% in New Delhi and Mangalore respectively) [20,21]. This could be attributed to greater awareness and stricter precautionary measures followed at these centres. However, the MRSA carrier rate among the non-exposed group (4%) is lower than the carrier rates of 11.1% and 18.1% obtained in two previous studies in India. This difference could be because our study included only a hundred participants of a specific age group (18y–23y), while the other studies surveyed entire populations [22,23]. The MRSA carrier rate in the exposed group is higher than that obtained in studies carried out in Canada (0.4%), Saudi Arabia (4.7%), Taiwan (5%) and France (6.2%). The MRSA carrier rate in the non-exposed group is higher than carrier rates in Saudi Arabia (1.3%) and Taiwan (3.5%), but lower than the carrier rate from a study in the USA (7.4%) [8,24–27].
The MRSA carrier rate of the exposed group was not significantly greater than that of the non-exposed group. This could be explained by contact of the nursing students with patients and others in the community, overcrowding, poor hygiene, and widespread over the counter use of antibiotics among the general population in India. Our result is the same as that obtained in a similar comparative study in Jordan [19], but differs from the results of two similar studies in Taiwan and Saudi Arabia, in which MRSA carrier rates in the hospital group were significantly more than in the non-hospital group [8,27].
The antibiotic resistance patterns of the MRSA isolates from the two study groups were examined. All isolates in both the groups showed susceptibility to netilmicin, linezolid, vancomycin and teicoplanin. This is in agreement with other Indian studies [21,23,28–30]. All isolates also showed susceptibility to tetracycline, and most to gentamicin. This, however, is in contrast to other Indian studies that showed high levels of resistance exhibited by MRSA strains towards the two antibiotics [23,28,29]. This could be attributed to the infrequent usage of tetracycline and gentamicin in our region. A study from Mangalore, a town in the same region of the country, also reported similar results [21].
Among the group exposed to hospital environment, most MRSA isolates demonstrated resistance towards cotrimoxazole, ciprofloxacin and erythromycin. This is in agreement with other Indian studies [23,28–30], and could be attributed to the frequent usage of these antimicrobials in a hospital setting.
Among the group not exposed to hospital environment, most MRSA isolates demonstrated resistance towards ciprofloxacin. The emergence of ciprofloxacin-resistant MRSA has been linked to the widespread use of ciprofloxacin for eradication of colonization and for the treatment of infections [3].
Overall, more MRSA isolates in the exposed group showed multi-drug resistance. This difference was, however, slight and could be explained as a response of the MRSA strains to the selection pressure created by their constant exposure to antibiotics used in a hospital environment.
All the MRSA isolates from both the study groups carried the lukS gene coding for pvl. It was originally thought that MRSA strains from the community characteristically carried genes coding for pvl [4]. However, recent studies show MRSA isolates from healthcare workers and patients may also express pvl [5–9]. All the MRSA isolates from both the study groups also expressed tsst-1 and hly a. The exotoxin sea was expressed by 63.6% (7/11) MRSA isolates in the exposed group, but by none in the non-exposed group (p = 0.0769; p > 0.05). The exotoxin seb was expressed by 90.9% (10/11) and 75% (3/4) MRSA isolates in the exposed and non-exposed groups respectively (p = 0.4762; p > 0.05). However, given the small number of isolates yielding these results for sea and seb in the two groups, these differences are not statistically significant.
The differences in carrier rates, antibiotic resistance patterns and expression of exotoxins between MRSA isolates from the two study groups are not significant, and pvl was found in all the MRSA isolates. These findings lead us to believe that the group exposed to hospital environment carried MRSA strains similar to those carried by the non-exposed group. Our study therefore suggests that healthcare workers could act as a link and transmit MRSA acquired from the community to patients under their care. This was impressed upon the nursing students in our study.
The findings of our study can be considered in the light of a recent mathematical model put forth to explain MRSA colonization in a hospital setting. This model suggests that community MRSA will become dominant in hospital settings due to a large community reservoir and increasing influx into the hospital of individuals who harbour these strains [31]. Our study appears to support this hypothesis of community MRSA infiltration in a hospital environment and suggests that healthcare workers play a role in its spread.
However, studies on a larger sample size in a controlled setting, with tracing of all contacts of the study participants, their follow-up over a period of time, and SCCmec and phage typing of the MRSA isolates would provide conclusive evidence.
Conclusion
The differences in carrier rates, antibiotic resistance patterns and expression of exotoxins between MRSA isolates from the two study groups were not significant, and pvl was found in all the MRSA isolates. These findings suggest that the nursing students carried MRSA strains similar to those carried by the non-exposed group and that they could act as a link and transmit MRSA acquired from the community to patients.
* Isolates with intermediate resistance were included with the resistant ones.
† Resistance to clindamycin was induced by erythromycin. These isolates showed a positive D-test.