Objectives: We determine the significant relation of HDL cholesterol and total cholesterol/HDL cholesterol between CETP I405V genotypes and activity of CETP.
CETP is an essential for transfer of cholesterol ester to the liver from peripheral tissues which facilitating its transfer to TG rich VLDL. Reduction activity of CETP I405V may associate with genotypes of CETP I405V. This study is undertaken to assess the presence and impact of CETP I405V genotype in our population.
Materials and Methods: In this study 100 acute myocardial infarction patients and 100 normal age & sex matched healthy individuals were included. Serum Lipid profile was estimated by using universal standard methods whereas CETP I405V genotype was studied by ARMS PCR.
Result: There is presence of CETP 405Val genotype both in patient as well as in control group. Results show that HDL cholesterol (p<0.0001) and ratio of total cholesterol/HDL cholesterol are significantly (p<0.0043) associated with Val/Val genotype. In addition to that the CETP I405V genotype is associated with inhibition of CETP activity with higher HDL-C level and decreased total cholesterol/HDL cholesterol ratio.
Conclusion: Our results show that the CETP I405V genotypes are very much significantly determinant of HDL cholesterol in patients with CHD.
Cholesteryl ester transfer protein (CETP), CETP I405V genotype,, High density lipoprotein cholesterol, Total cholesterol, Triacylglyceride
Introduction
High density lipoprotein (HDL) cholesterol plays a central role in the transport of cholesterol from peripheral tissues to the liver. This process of reverse cholesterol transport [1,2] explains the major antiatherogenic mode of action of this lipoprotein fraction. This process involves the esterification of free cholesterol within the HDL fraction and its subsequent transfer by Cholesteryl Ester Transfer Protein (CETP) to TG-rich lipoproteins [3].
The CETP gene is one of the major genes affecting HDL metabolism; the gene codes for CETP, a protein responsible for the transfer of cholesteryl ester (CE) from HDL to apolipoprotein B-containing particles, particularly very low-density lipoprotein (VLDL), in exchange for triacylglyceride [4]. Human CETP gene consists of 16 exons located on chromosome 16 adjacent to lecithin cholesterol acyl transferase (LCAT) gene [5]. Some genotypes like the R451Q [6] and A373P [7] are associated with high plasma CETP activity, low HDL-C. The other genetic alterations such as Int14A, D422G genotypes [8–9] and I405Vgenotype [10] have been identified as cause of low or deficient CETP activity and elevated levels of HDL-C [11].
There are scanty reports of CETP genotype studied and its correlation with lipid profile in the Indian population. We studied CETP genotype and their correlation with lipid levels in acute myocardial infarction patients and healthy controls from Western Maharashtra region, India.
Materials and Methods
Subjects
The present case-control study was performed in the Department of Biochemistry, Government Medical College, Miraj, India. Total 100 acute myocardial infarction (AMI) patients, out of which 67 were males and 33 were females, within age ranging from 19 to 88 years and 100 normal age and sex matched healthy individuals as controls were included. All control samples were free from diseases like diabetes mellitus, hypertension, coronary artery disease, atherosclerosis, acute or chronic renal or liver disease, no history of thyroid impairment and smoking and/or tobacco chewing.
The diagnosis of acute myocardial infarction was done by the physicians of General Hospital, Sangli and General Hospital Miraj; based on clinical history, electrocardiogram, and relevant biochemical parameters. Fasting blood sample of patients and controls were collected and analysed. The protocol was approved by the Institutional Ethical committee and subjects provided written informed consent.
Methods
The fasting blood samples of control and MI subjects were collected; for MI subjects sample was collected within three days of diagnosis of the disease. Serum samples were analysed for lipid profile and EDTA samples were used for extraction of DNA.
Serum Total Cholesterol level was estimated by enzymatic method [12]. For HDL cholesterol estimation, Chylomicrons, VLDL cholesterol and low density lipoprotein (LDL) cholesterol fractions in serum was separated by precipitating with phosphotungstic acid and magnesium chloride. After centrifugation the cholesterol in the HDL fraction, which remains in the supernatant, was assayed with enzymatic cholesterol method [12]. LDL cholesterol level was calculated using Friedwald’s formula [13]. Serum Triacylglyceride was determined by Glycerol phosphate oxidase-peroxidase method [14].
Genetic analysis
EDTA blood samples were used for DNA extraction by salting out method [15]. Genotypes for CETP I405V for all above subjects and controls were determined by Amplification Refractory Mutation System PCR [16]. The mutant and wild specific ARMS primers, control primers, and common primer used to diagnose CETP I405V genotype in this study are listed in [Table/Fig-1].
The mutation-specific ARMS primers, control primers, and common primer for CETP I405V genotype
| S. No | Mutation/wild specific/common/ control primer | Primer sequence (5’–3’) | Length of PCR product (bp) |
|---|
| 1 | CETP 405 Wild | GCAGAGCAGCTCCGAGTTCA | 646 bp |
| 2 | CETP 405 Mut | CAGAGCAGCTCCGAGACCG |
| 3 | CETP 405 Com | ATGTTCAAAAGGAAAGCACAGTC | |
| 4 | CONTROL FOR | CCCACCTTCCCCTCTCTCCAG GCAAATGGG | 360 bp |
| 5 | CONTROL RES | GGGCCTCAGTCCCAACATGGC TAAGAGGTG |
A common primer was designed that paired with CETP I405V giving a product of 646 base pairs (bp). A 360-bp fragment of the α1-antitrypsin gene was co-amplified to function as an internal control. For each reaction there were total four primers used. For CETP I405V genotypes wild/mutant primer, common primer and two control primers were used.
The PCR was carried out in a total volume of 25 μl of reaction mixture, containing 50 ng of genomic DNA; 12.5 μl 2X master mix giving final concentration 200 mmol/L of deoxynucleotides triphosphates; 10 mmol/L Tris-HCL, pH 8.8; 1.5 mmol/L MgCl2; 50 mmol/L KCl; 2.5 U of Taq DNA Polymerase; and then 8 nmol/L of control primers, and 0.8 mmol/L ARMS common primer. Reaction mixture wild type contained, in addition to the above, 0.2 ~ 0.5 pmol / L CETP I405V wild primer. Similarly, reaction mixture mutant type contained, in addition to the above, 0.2 ~ 0.5 pmol / L CETP I405V mutant primer. Amplification was performed with a Quanta Biotech DNA thermocycler. Each PCR run had negative controls. Cycling was carried out on the thermal cycler with initial denaturation at 95°C for 5 min, 32 cycles with denaturation at 94°C for 50 s, annealing at 66 °C for 45 s, and extension at 72°C for 60 s, followed by a final cycle of extension at 72°C for 5 min. Total 10 μl of amplified product was mixed with 2.0 μl of gel loading dye and fragments separated in 2% agarose gel that contained 0.1 mg/L ethidium bromide for visualization. The samples were electrophoresed for 1 hour at 100 volts, using 1x TAE (Tris-Acetate-EDTA) running buffer The DNA fragments were visualized by staining with Ethidium bromide (EtBr). This dye binds to DNA and fluoresces under ultraviolet radiation.
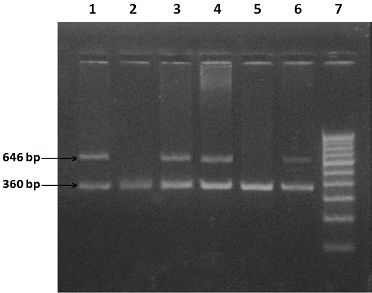
Amplification products of the CETP I405V genotypes with modified ARMS on agarose gel electrophoresis are shown in [Table/Fig-2].
Photograph showing electrophoresis of ARMS-PCR for CETP I405V genotype
1, 3 & 4- Samples positive for V genotype of CETP I405V
2. Sample negative for V genotype of CETP I405V
5. Negative control for genotype CETP I405V
6. Positive control for genotype CETP I405V
7. 100 bp DNA marker ladder
Statistical Analysis
Within patient group, Hardy–Weinberg equilibrium was tested by means of chi-square analysis. Throughout, independent samples test-test was used and p-value of <0.05 was interpreted as indicating a statistically significant difference. All statistical analyses were carried out with MEDCALC software.
Results
Total 200 samples were studied, out of which 100 were acute myocardial infarction patients and 100 were control subjects. [Table/Fig-3] shows serum lipid profile in study and control groups. The total cholesterol level was 220.11 ± 55.70 mg/dl in patients and 170.92 ± 22.45 mg/dl in controls, which is found to be statistically significant (p<0.0001). The HDL cholesterol level was 37.83 ± 12.58 mg/dl in patients and 49.14 ± 10.50 mg/dl in controls, which is significantly (p<0.0001) decreased in patients than in controls. Total cholesterol/HDL cholesterol ratio was 6.48 ± 2.88 in patients and 3.65 ± 0.98 in controls which is significantly increased (p<0.0001) in patients than in controls. Mean values for serum LDL cholesterol was 158.22 ± 58.22 in patients and 103.36 ± 25.40 in controls which is significantly higher (p<0.0001) among myocardial infarction patients than the controls. The VLDL cholesterol level was 24.15 ± 12.03 mg/dl in patients and 18.41 ± 5.67 mg/dl in controls, which is found to be statistically significant (p<0.0001). The triacylglycerol level was 120.76 ± 60.16 mg/dl in patients and 92.07 ± 28.39mg/dl in controls, which is significantly (p<0.0001) increased in myocardial patients than in controls.
Biochemical parameters in MI patients and controls
| Biochemical parameters | Patients (n=100) | Controls (n=100) | p-value |
|---|
| Total Cholesterol (mg/dl) | 220.21 ± 55.70 | 170.92 ± 22.45 | p < 0.0001* |
| HDL Cholesterol (mg/dl) | 37.83 ± 12.58 | 49.14 ± 10.50 | p < 0.0001* |
| Total Cholesterol /HDL Cholesterol | 6.48 ± 2.88 | 3.65 ± 0.98 | p < 0.0001* |
| LDL Cholesterol (mg/dl) | 158.22 ± 58.22 | 103.36 ± 25.40 | p < 0.0001* |
| VLDL Cholesterol (mg/dl) | 24.15 ± 12.03 | 18.41 ± 5.67 | p < 0.0001* |
| Triacylglycerol (mg/dl) | 120.76 ± 60.16 | 92.07 ± 28.39 | p < 0.0001* |
| ’*’ Significant |
The observed frequencies of genotypes were in Hardy–Weinberg equilibrium with Chi-square p-value 0.4795. Frequency Distribution of Ile 405 Val genotype of CETP gene in myocardial infarction patients and controls were as depicted in [Table/Fig-4]. The frequencies of CETP I405V genotype in patient group were Ile/Ile (32%), Ile/Val (46%) and Val/Val (22%), while those in control group were Il/Ile (38%), Ile/Val (44%) and Val/Val (18%), which is not significant with patients compared to controls.
Frequency distribution of Ile 405Val genotype of CETP in MI patient & controls
| Genotypes | Patients (n=100) | Controls (n=100) | p-value |
|---|
| Ile/Ile | 32 % | 38 % | 0.4585# |
| Ile/Val | 46 % | 44 % | 0.8870# |
| Val/Val | 22 % | 18 % | 0.5959# |
| ’#’ not significant., X2 test p-value of patients is 0.4795 |
We also compare the lipid levels in group having Ile/Ile genotype with Val/Val genotype in myocardial infarction patients, as shown in [Table/Fig-5]. The group with Ile/Ile genotype had significantly Lower HDL cholesterol (p<0.0001), and higher total cholesterol/HDL cholesterol ratio (p=0.0043) and no significant difference found in Total cholesterol (p=0.5413), LDL cholesterol (p=0.6166), VLDL cholesterol (p=0.6681), plasma triacylglyceride (p=0.6681) when compared to the Val/Val genotype of CETP I405V.
Comparison of biochemical parameters of CETP I405V genotype in study subjects with Ile/Ile group with Val/Val group in MI patients
| Biochemical parameter | Ile/ Ile (n=32) | Val/ Val (n=22) | p-value |
|---|
| Total Cholesterol mg/dl | 216.53 ± 57.38 | 225.74 ± 48.78 | p = 0.5413# |
| HDL Cholesterol mg/dl | 32.66 ± 8.67 | 48.50 ± 15.51 | p < 0.0001* |
| Total cholesterol/HDL cholesterol | 7.06 ± 2.43 | 5.15 ± 2.11 | p = 0.0043* |
| LDL Cholesterol mg/dl | 160.26 ± 58.81 | 152.46 ± 51.30 | p = 0.6166# |
| VLDL Cholesterol mg/dl | 23.61 ± 7.63 | 24.78 ± 12.21 | p= 0.6681# |
| Triglycerides mg/dl | 118.07 ± 38.13 | 123.88 ± 61.04 | p= 0.6681# |
| ’*’ significant. ’#’ not significant |
Discussion
CETP is essential in the reverse cholesterol transport pathway, the only route to eliminate excess cholesterol from body. After HDL particles accept cholesterol from extra hepatic tissues, CETP facilitates the transfer of cholesteryl ester onto triglyceride rich lipoproteins as part of the reverse cholesterol transport pathway, ultimately leading to cholesterol excretion by the liver [17,18].
Freeman et al., [19] initially reported an inverse association between HDL and CETP activity in highly selected population samples for low and high HDL cholesterol levels. CETP I405V genotype is the most important inherited trait modulating CETP activity and serum HDL cholesterol. We examined the effect of CETP I405V genotype on lipid level in a south west Maharashtra population originating from western India.
In the present study, we found significantly increased levels of serum cholesterol, triacylglyceride, LDL cholesterol, VLDL cholesterol and total cholesterol/HDL cholesterol ratio in myocardial infarction patients when compared to controls, while HDL cholesterol was significantly decreased in MI patients. We found 22 myocardial infarction patients and 18 normal healthy controls with Val/ Val genotype. There was no significant difference in frequency of CETP I405V in patients when compared with controls. This genotype is shown to be associated with a reduction in CETP activity, and increased level of HDL cholesterol [20–23].
We have studied a relationship between CETP I405V genotype and lipid profile. The lipid levels were compared in patients of myocardial infarction on the basis of the genotype present i.e. in between Ile/Ile and Val/Val genotypes. HDL cholesterol was significantly higher in patients carrying the Val/Val genotype than with the Ile/Ile genotype, and lower total cholesterol/HDL cholesterol ratio in Val/Val genotype than patients with Ile/Ile genotype. There was no significant difference found in total cholesterol, LDL cholesterol, VLDL cholesterol and triacylglycerol in patients with Ile/Ile & Val/Val genotype of CETP I405V.
From the above results we conclude that in myocardial infarction the presence of Val/Val is found to be associated with increased HDL cholesterol, and decreased atherogenic ratio total cholesterol/HDL cholesterol was found in patients with Val/Val genotype. The increase in HDL cholesterol may not be beneficial as per Todur SP et al.,[24] as they found no association between CETP I405V genotype and CAD, but it is an important determinant of HDL cholesterol.
CETP deficiency due to genetic defect in CETP is an autosomal recessive phenomenon caused by homozygous or heterozygous CETP genotypes [25]. The CETP I405V genotype affect level of HDL cholesterol and apolipoprotein A-I, the major protein in HDL particles. Complete CETP deficiency leads to massively elevated levels of HDL cholesterol and apolipoprotein A-I, [26–29] and our results are in accordance with previous studies, who demonstrated dysfunction of CETP due to CETP I405V genotype which leads reduced reverse cholesterol transport and increase in HDL cholesterol level and decreased risk of atherosclerosis [30,31].
In an analysis of the CETP I405V genotype of the Copenhagen City Heart Study by Agerhoim-Larsen B [32]. Found that women for the presence of valine at position 405 of CETP gene had increased levels of HDL cholesterol and total cholesterol/HDL cholesterol ratio was significantly lower with Val/Val genotype compared to Ile/Ile genotype in women having anti-atherogenic effect of Val/Val genotype. And no significant difference was found in men for total cholesterol/HDL cholesterol in Val/Val genotype and Ile/Ile genotype showing no risk of atherogenecity.
Bruce C et al., [25] found significantly lower CETP in Val/ Val genotype than Ile/ Ile and Ile/ Val genotype and significantly higher HDL cholesterol than Ile/ Ile and Ile/Val genotype, and showed the CHD risk was higher due to hypertriglyceridemia with the Val/Val genotype than in those with the Ile/Val or Ile/Ile genotype of CETP I405V. Pallaud C et al., [33] found no association between the CETP I405V genotype and either HDL cholesterol levels or CHD risk in the men.
Conclusion
Data demonstrate that variation at the CETP gene locus is a significant determinant of HDL-C levels, CETP activity, and probably a genetic risk factor for CHD in the study population
[1]. Bruce C, Tall AR, Cholesteryl ester transfer proteins, reverse cholesterol transport, and atherosclerosisCurr Opin Lipidol 1995 6:306-11. [Google Scholar]
[2]. Fielding CJ, Fielding PE, Molecular physiology of reverse cholesterol transportJ Lipid Res 1995 36:211-28. [Google Scholar]
[3]. Tall AR, Plasma cholesteryl ester transfer proteinJ Lipid Res 1993 34:1255-74. [Google Scholar]
[4]. Chapman MJ, Le Goff W, Guerin M, Kontush A, Cholesteryl ester transfer protein: at the heart of the action of lipid-modulating therapy with statins, fibrates, niacin, and cholesteryl ester transfer protein inhibitorsEur Heart J 2010 31:149-64. [Google Scholar]
[5]. Kakko S, Tamminen M, Kesaniemi YA, Savolainen MJ, R451Q mutation in the cholesteryl ester transfer protein (CETP) gene is associated with high plasma CETP activityAtherosclerosis 1998 136(2):233-40. [Google Scholar]
[6]. Agerholm-Larsen B, Tybjaerg-Hansen A, Schnohr P, Steffensen R, Nordestgaard BG, Common cholesteryl ester transfer protein mutations, decreased HDL cholesterol, and possible decreased risk of ischemic heart disease: The Copenhagen City Heart StudyCirculation 2000 102(18):2197-203. [Google Scholar]
[7]. Zhong S, Sharp DS, Grove JS, Bruce C, Yano K, Curb JD, Increased coronary heart disease in Japanese-American men with mutation in the cholesteryl ester transfer protein gene despite increased HDL levelsJ Clin Invest 1996 97(12):2917-23. [Google Scholar]
[8]. Wang J, Qiang H, Chen D, Zhang C, Zhuang Y, CETP gene genotype (D442G) increases low-density lipoprotein particle size in patients with coronary heart diseaseClin Chim Acta 2002 322(1-2):85-90. [Google Scholar]
[9]. Zhuang Y, Wang J, Qiang H, Li Y, Liu X, Li L, Cholesteryl ester transfer protein levels and gene deficiency in Chinese patients with cardio-cerebrovascular diseasesChin Med J (Engl) 2002 115(3):371-4. [Google Scholar]
[10]. Kakko S, Tamminen M, Paivansalo M, Kauma H, Rantala AO, Lilja M, Cholesteryl ester transfer protein gene genotypes are associated with carotid atherosclerosis in menEur J Clin Invest 2000 30(1):18-25. [Google Scholar]
[11]. Thompson JF, Lira ME, Durham LK, Clark RW, Bamberger MJ, Milos PM, Genotypes in the CETP gene and association with CETP mass and HDL levelsAtherosclerosis 2003 167(2):195-204. [Google Scholar]
[12]. Allain CC, Poon LS, Chan CS, Richmond W, Fu PC, Enzymatic determination of total serum cholesterolClin Chem 1974 20(4):470-5. [Google Scholar]
[13]. Friedewald WT, Levy RI, Fredrickson DS, Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifugeClin Chem 1972 18:499-502. [Google Scholar]
[14]. Bucolo G, David H, Quantitative determination of serum triglycerides by the use of enzymesClin Chem 1973 19(5):476-82. [Google Scholar]
[15]. Miller SA, Dykes DD, Polesky HF, A simple salting out procedure for extracting DNA from human nucleated cellsNucleic Acids Research 1988 16(3):1215 [Google Scholar]
[16]. Dai-feng Z, Wang-wei C, Mei-ling Y, Zhen Z, Genotype of CETP gene in the healthy population of Han and Li nationality in Hainan provinceBasic & clinical Medicine 2001 30(7):754-58. [Google Scholar]
[17]. Barter PJ, Rye KA, High density lipoproteins and coronary heart diseaseAtherosclerosis 1996 121:1-12. [Google Scholar]
[18]. Breslow JL, Familial disorders of high-density lipoprotein metabolism. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Stanbury JB, Wyngaarden JB, Frederickson DS, edsThe Metabolic and Molecular Bases of Inherited Disease 1995 7th ednNew York, NYMcGraw-Hill:2031-52. [Google Scholar]
[19]. Freeman DJ, Packard CJ, Shepherd J, Gaffney D, Polymorphisms in the gene coding for cholesteryl ester transfer protein are related to plasma high-density lipoprotein cholesterol and transfer protein activityClin Sci 1990 79:575-81. [Google Scholar]
[20]. Okumura K, Matsui H, Kamiya H, Saburi Y, Hayashi K, Hayakawa T, Differential effect of two common genotypes in the cholesteryl ester transfer protein gene on low-density lipoprotein particle sizeAtherosclerosis 2002 16:425-31. [Google Scholar]
[21]. Barzilai N, Atzmon G, Schechter C, Schaefer EJ, Cupples AL, Lipton R, Unique lipoprotein phenotype and genotype associated with exceptional longevityJ Am Med Assoc 2003 290:2030-40. [Google Scholar]
[22]. Boekholdt MS, Kuivenhoven JA, Kees HG, Wouter JJ, Kastelein JJP, Van TA, CETP gene variation: relation to lipid parameters and cardiovascular riskCurr Opin Lipidol 2004 15:393-8. [Google Scholar]
[23]. Brousseau ME, Schaefer EJ, Wolfe ML, Bloedon LT, Digenio AG, Clark RW, Effects of an inhibitor of cholesteryl ester transfer protein on HDL cholesterolN Engl J Med 2004 350:1505-15. [Google Scholar]
[24]. Todur SP, Tester FA, Association of CETP and LIPC Gene Polymorphisms with HDL and LDL Sub-fraction Levels in a Group of Indian Subjects: A Cross-Sectional StudyIJCB 2013 28(2):116-23. [Google Scholar]
[25]. Bruce C, Sharp DS, Tall AR, Relationship of HDL and coronary heart disease to a common amino acid genotype in the cholesteryl ester transfer protein in men with and without hypertriglyceridemiaJ Lipid Res 1998 39:1071-78. [Google Scholar]
[26]. Yamashita S, Hui DY, Sprecher DL, Matsuzawa Y, Sakai N, Tarui Total deficiency of plasma cholesteryl ester transfer protein in subjects homozygous and heterozygous for the intron 14 splicing defectBiochem Biophys Res Commun 1990 170:1346-51. [Google Scholar]
[27]. Makita H, Tsuji M, Furuya Y, Tsuchihashi K, Akita H, Chiba H, A family with complete deficiency of plasma cholesteryl ester transfer protein activitiesIntern Med 1994 33:432-6. [Google Scholar]
[28]. Matsunaga A, Araki K, Moriyama K, Handa K, Arakawa F, Nishi K, Detection of a point genotype in cholesteryl ester transfer protein gene by polymerase chain reaction-mediated site-directed mutagenesisBiochim Biophys Acta 1993 1166:131-4. [Google Scholar]
[29]. Hirano K, Yamashita S, Nakajima N, Arai T, Maruyama T, Yoshida Y, Genetic cholesteryl ester transfer protein deficiency is extremely frequent in the Omagari area of Japan: marked hyperalphalipoproteinemia caused by CETP gene genotype is not associated with longevityArterioscler Thromb Vasc Biol 1997 17:1053-9. [Google Scholar]
[30]. Kuivenhoven JA, de Knijff P, Boer JM, Smalheer HA, Botma GJ, Seidell Heterogeneity at the CETP gene locus: influence on plasma CETP concentrations and HDL cholesterol levelsArterioscler Thromb Vasc Biol 1997 17:560-8. [Google Scholar]
[31]. Gudnason V, Kakko S, Nicaud V, Savolainen MJ, Kesaniemi YA, Tahvanainen E, Cholesteryl ester transfer protein gene effect on CETP activity and plasma high-density lipoprotein in European populationsEur J Clin Invest 1999 29:116-28. [Google Scholar]
[32]. Agerholm-Larsen B, Nordestgaard BG, Steffensen R, Jensen G, Tybjaerg-Hansen A, Elevated HDL cholesterol is a risk factor for ischemic heart disease in white women when caused by a common mutation in the cholesteryl ester transfer protein geneCirculation 2000 101:1907-12. [Google Scholar]
[33]. Pallaud C, Gueguen R, Sass C, Grow M, Cheng S, Siest G, Genetic influences on lipid metabolism trait variability within the Stanislas CohortJ Lipid Res 2001 42:1879-90. [Google Scholar]