Objective: To detect genes encoding carbapenem resistance in urinary isolates of Escherichia coli recovered from hospitalized patients in tertiary care centre in Pune, India.
Methods: From Jan 2012 to Dec 2012, a total of 300 consecutive non-duplicate (one isolate per patient) clinical isolates of Escherichia coli were recovered from urine cultures of hospitalized patients including hospital acquired infection cases admitted to the medical and surgical intensive care units. Polymerase chain reaction (PCR) assays and sequencing was used to determine the presence of beta-lactamase encoding genes. Conjugation experiments were performed to determine the transferability of beta-lactamase.
Results: All the isolates were completely resistant to the second and third generation cephalosporins tested as well as carbapenems. All the isolates showed 100% susceptibility to tigecycline and colistin in vitro. Conjugation experiments demonstrated that blaNDM-1 was transferable via plasmid. All the isolates showed presence of blaNDM-1 and co-association of blaOXA-48 was 25/45(55%) of the isolates. Repetitive element based PCR (REP PCR), Enterobacterial Repetitive Intergenic Consensus (ERIC PCR) and Randomly Amplified Polymorphic DNA (RAPD) revealed a diversity of six clonal types among E.coli isolates.
Conclusion: Co-production of NDM-1with OXA-48 in urinary isolates of E. coli was detected for the first time in India. Transmission of plasmid carrying these resistant genes to other members of Enterobacteriaceae will increase incidence of multidrug resistance. Early detection of these genes will help in prevention and adequate infection control by limiting the spread of these organisms.
Introduction
Beta-lactam antibiotics are one of the most frequently used antimicrobials in hospital settings used to treat infections caused by Gram negative bacteria. Escherichia coli is one of the most common pathogen of Enterobacteriaceae family responsible for nosocomial infections. However of lately due to the presence of extended-spectrum beta- lactamase (ESBL) and AmpC enzymes in these Gram negative bacilli, carbapenems have become the drug of choice to treat such infections. Incidence of multi drug resistance in organisms is increasing due to dissemination of resistance determinant genes mediated by gene cassettes in integrons, plasmids and transposons. Resistance to carbapenems due to carbapenemase production poses serious challenges in the treatment of such infections with pan-resistant phenotypes [1]. In this study molecular detection of resistant genes along with plasmid replicon typing of carbapenem resistant urinary isolates of E.coli was carried out and genes encoding both NDM-1 and OXA-48 in E.coli were detected.
Materials and Methods
The Bacterial Clinical Isolates
The study was conducted after obtaining due approval from the institutional ethical committee. From Jan 2012 to Dec 2012, a total of 300 consecutive non-duplicate (one isolate per patient) clinical isolates of E.coli recovered from urine culture of hospitalized patients admitted to the medical and surgical intensive care units in 1000 bedded tertiary care hospital in Pune, India, were included in the study. Collection of urine sample was done using strict aseptic precautions and was immediately processed without any delay. Urine culture was carried out on Cysteine Lactose Electrolyte Deficient (CLED) agar medium using calibrated standard loop. Bacterial identification was performed by routine conventional microbial culture and biochemical tests using standard recommended techniques [2]. The organisms were identified up to the species level using VITEK-GNI cards (bioMérieux, Marcy l’Etoile, France).
Antimicrobial Susceptibility Testing and MIC Determination
Antibiotic sensitivity test was performed by standard Kirby Bauer disc diffusion technique as per the guidelines of the Clinical Laboratory Standards Institute (CLSI) with commercially available discs (Hi Media, Mumbai, India) on Mueller Hinton agar plates [3].The antibiotics tested were as follows (potency in μg/disc): piperacillin (100), ticarcillin (75), piperacillin-tazobactam (100/10), ticarcillin-clavulanic acid (75/10), ceftazidime (30), cefotaxime (30), cefepime (30),cefoxitin (30), ceftriaxone (30), aztreonam (30), imipenem (10), meropenem (10), ertapenem (10), gentamicin (10), tobramycin (10), amikacin (30), netilmicin (30), ciprofloxacin (5), levofloxacin (5), lomefloxacin (10) and ofloxacin (5). P. aeruginosa ATCC 27853, E.coli ATCC 25922, E. coli ATCC 35218 and K. pneumoniae ATCC 700603 were used as quality control strains. Minimum inhibitory concentrations (MIC) of antibiotics were determined by VITEK-2 AST-GN25 and AST-GN280 susceptibility cards in accordance with the Clinical and Laboratory Standards Institute (CLSI) recommendations and manufacturers’ instructions, except tigecycline and colistin, for which the 2012 European Committee on Antimicrobial Susceptibility Testing break points were used [3,4].MICs were further determined by the E-test (bioMérieux, Marcy l’Etoile, France).
Phenotypic Screening for the Carbapenemase Production
E.coli isolates with reduced susceptibility to meropenem and imipenem (diameter of zones of inhibition ≤13mm) by disc diffusion method were screened for the production of carbapenemase. The phenotypic detection of the carbapenemase production was performed by the modified Hodge test by using a meropenem disc (10 μg) as per CLSI guidelines [3]. For MHT K. pneumoniae ATCC BAA-1705 and BAA-1706 were used as positive and negative controls, respectively. The screening of metallo-beta-lactamase production was performed by the double-disc synergy tests (DDST) and combined-disc synergy test (CDST) as described previously [5,6]. K. pneumoniae ATCC BAA-2146 and P. aeruginosa ATCC 27853 were used as positive and negative controls, respectively. MBL (IP/IPI) E-test was carried out to detect MBL as per manufacturer’s instructions.
Molecular detection of the Beta-lactamase genes
DNA was extracted using the spin column method (QIAGEN; GmbH, Hilden, Germany) as per manufacturer’s instructions. PCR-based detection of beta lactamase (ESBL) genes (blaCTXM, blaSHV, blaTEM and blaOXA), Ambler class B MBLs (blaIMP, blaVIM, blaSPM, blaGIM, blaSIM and blaNDM-1), Ambler class D (blaOXA-23, blaOXA-24 and blaOXA48) and for serine carbapenemases (blaKPC, blaGES and blaNMC) were carried out on the isolates by using Gene Amp 9700 PCR System (Applied Biosystems, Singapore) [7]. PCR products were run on 1.5% agarose gel, stained with ethidium bromide visualized under UV light and photographed. The amplicons were purified using QIAquick PCR purification kit (QIAGEN; GmbH, Hilden, Germany).
DNA Sequencing and Sequence Analysis
Automated sequencing was performed on an ABI 3730XL DNA analyzer using the Big Dye system (Applied Biosystems Foster City, CA, USA). Sequences were compared with known sequences using the BLAST facility (http://blast.ncbi.nlm.nih.gov).
Conjugation Experiments
Transfer of resistance genes by conjugation was assayed by mating experiments in Luria–Bertani broth using E. coli urinary isolates (Parental strains) as donors and an azide-resistant E. coli J53 as the recipient strain using 1:10 ratio. The transconjugants were selected on Luria–Bertani agar with selection based on growth on agar in the presence of ceftazidime (30 µg/ml) and sodium azide (100 µg/ml). Plasmids were separated by co-electrophoresis on horizontal 0.5% agarose gel at 50 volts for 3 Hrs. The size of the plasmids were compared by co electrophoresis with plasmid of known sizes from E.coli (V517 and 39R861). Bands were visualized with UV transilluminator after staining with 0.05% ethidium bromide.
Strain Molecular Typing
Repetitive element based PCR (REP-PCR) and Enterobacterial Repetitive Intergenic Consensus (ERIC-PCR) assays were performed as described to rapidly characterize blaNDM-1 positive E. coli strains recovered from patients [8]. Randomly Amplified Polymorphic DNA (RAPD) analysis was also carried out to detect relatedness of the strains [9].
Plasmid Analysis
Plasmid from the parental strains and their transconjugants was extracted by using Qiagen plasmid mini kit (GmbH, Hilden, Germany) as per manufacturer’s Instructions. Extracted plasmid DNA were subjected to plasmid based replicon incompatibility (Inc) typing by using 18 pairs of primers to perform five multiplex and three single PCRs which recognized F, FIA, FIB, FIC, B/O, X, Y, N, P, W, T, A/C, HI1, HI2, I1-Ic, L/M, K and FII replicons as described previously [10]. Plasmid replicons were determined for the ESBL as well as carbapenemase producing clinical isolates.
Results
Out of total 300 clinical urinary isolates of Escherichia coli, 45 were found to be carbapenem (imipenem, meropenem and ertapenem) resistant by the disk diffusion test and by e-test. These isolates showed resistance to other beta lactam antibiotics, aminoglycosides and quinolones tested. Carbapenemase production was confirmed by Modified Hodge test. Production of MBL was confirmed by positive DDST, CDST and MBL (IP/IPI) E-test method. All 45 carbapenem resistant isolates found to be positive for blaNDM-1 [Table/Fig-1] and 25 among these isolates found to be positive for blaOXA-48 [Table/Fig-2]. Antibiogram of these 25 isolates are depicted in [Table/Fig-3].The PCR results were validated by sequencing. Overall blaCTX-M-15 was the commonest genotype 38/45 (84%) followed by blaTEM32/45(71%), blaSHV28/45(62%) and blaOXA 19/45(42%) either alone or in combination [Table/Fig-4].Based on sequencing it was confirmed that blaTEM-1, blaSHV-5, blaSHV-11, blaSHV-12, blaSHV-28, blaCTX-M-15, blaOXA-1, blaOXA-2, blaOXA-9 and blaOXA-10 are the dominant ESBLs among these carbapenem resistant strains in our study. Conjugation experiments indicated that blaNDM-1 was transferable via a plasmid along with other beta lactamase genes carried on other plasmids. Plasmid profiling of the isolates showed that blaNDM-1 was carried on plasmids ranging in sizes from 70 to120 kb and blaOXA-48 was carried on 50 kb size plasmids.
REP-PCR, ERIC-PCR and RAPD assays confirmed presence of 6 clones as per banding pattern. Plasmids purified from the clinical isolates were typed by PCR based replicon typing. IncFIA, IncFIB, IncFIC replicons were associated with blaTEM-1.Majority of blaSHV showed association with multiple replicons (either IncFII, IncFIB or IncFIA, IncFIB), three isolates showed single replicon association (IncFIC). The blaNDM-1gene in E.coli was located on IncA/C plasmid. The blaOXA-48 carried on plasmids belonging to Inc L/M replicons.BlaCTX-M-15 was associated with multiple replicons of plasmid (IncFIA, IncFIB). The blaOXA identified on plasmids was associated with IncP and IncW replicons [Table/Fig-5].
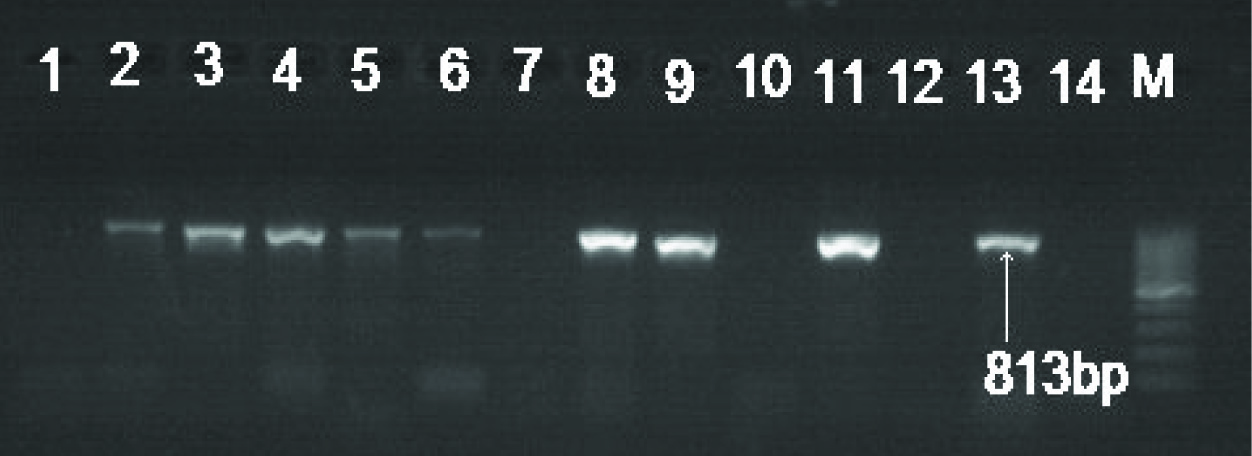
Electrophoretic banding pattern on 1.5 % agarose gel showing PCR amplified products of blaNDM-1 (813 bp). Lanes 1-12 urinary E. coli isolates positive for NDM-1, Lane 13- positive control for NDM-1, Lane 14- negative Control and M represents; 100 bp DNA ladder used as the molecular size standard
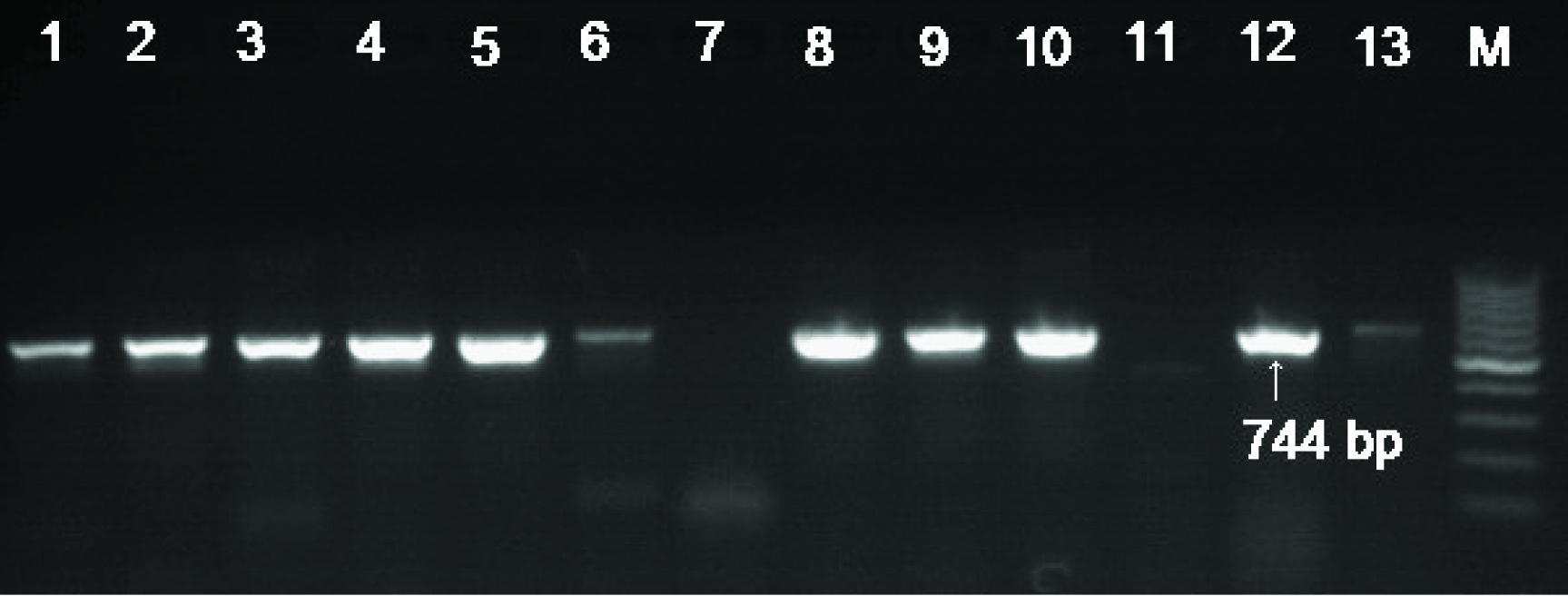
Electrophoretic banding pattern on 1.5 % agarose gel showing PCR amplified products of blaoxa48 (744 bp). Lanes 1-6 and 9-13urinary E. coli isolates positive for Oxa48, Lane 7- negative Control, Lane 8- in-house positive control for Oxa48 and M represents; 100 bp DNA ladder used as the molecular size standard
Showing MICs of urinary E. coli isolates against antibiotic used co-producing NDM-1 and OXA-48 carbapenemases (n=25)
| Antibiotic used | MIC μg/ml |
| amikacin [AMK] | 64 |
| ampicillin[AMP] | 128 |
| cefepime [CPM | 64 |
| ceftazidime [CAZ] | 128 |
| cefuroxime [CFR] | 64 |
| ciprofloxacin[CIP] | 4 |
| Cefotaxime[CTX] | 128 |
| ertapenem[ETP] | 8 |
| gentamicin [GEN] | 16 |
| imipenem [IPM] | 64 |
| levofloxacin [LEV] | 8 |
| meropenem [MEM] | 64 |
| moxifloxacin[MXF] | 8 |
| piperacillin[PIP] | 128 |
| tobramycin [TOB] | 16 |
| colistin [CST] | <0.5 |
| tigecycline [TGC] | <2 |
Showing distribution of Beta Lactamase genes in multidrug resistant urinary E. coli isolates (n=45)
| Name of gene | Present | Percentage |
| NDM-1 | 45 | 100% |
| OXA 48 | 25 | 55% |
| CTX-M-15 | 38 | 84% |
| TEM-1 | 32 | 71% |
| SHV type ESBL SHV5, SHV11, SHV12, SHV28 | 28 | 62% |
| OXA type ESBL OXA-1, OXA-2, OXA-9, OXA10 | 19 | 42% |
Showing replicon typing in multidrug resistant E. coli isolates co-producing NDM-1 and OXA-48 carbapenemases
| I.D | TEM | SHV | CTX-M | OXA | OXA- 48 | NDM-1 |
| UC10 | FIB | FII,FIB | FIA,FIB | P | L/M | A/C |
| UC53 | FIC | NG | FIA,FIB | NG | L/M | A/C |
| UC66 | FIB | FIA,FIB | FIA,FIB | P | L/M | A/C |
| UC251 | FIC | FII,FIB | FIA,FIB | NG | L/M | A/C |
| UC320 | FIA | FII,FIB | FIA,FIB | P | L/M | A/C |
| UC405 | FIB | NG | FIA,FIB | W | L/M | A/C |
| UC480 | FIA | FIA,FIB | FIA,FIB | NG | L/M | A/C |
| UC1387 | FIC | FIC | FIA,FIB | NG | L/M | A/C |
| UC1445 | NG | FIC | FIA,FIB | P | L/M | A/C |
| UC1632 | NG | FIA,FIB | FIA,FIB | NG | L/M | A/C |
| UC1862 | FIB | FII,FIB | FIA,FIB | NG | L/M | A/C |
| UC1880 | FIC | NG | FIA,FIB | W | L/M | A/C |
| UC2044 | FIC | FII,FIB | FIA,FIB | W | L/M | A/C |
| UC2330 | FIA | FIA,FIB | FIA,FIB | NG | L/M | A/C |
| UC2454 | NG | FIC | FIA,FIB | NG | L/M | A/C |
| UC2632 | FIB | FII,FIB | FIA,FIB | W | L/M | A/C |
| UC2778 | FIC | FII,FIB | FIA,FIB | P | L/M | A/C |
| UC2981 | FIA | FII,FIB | FIA,FIB | P | L/M | A/C |
| UC3028 | FIB | FII,FIB | FIA,FIB | NG | L/M | A/C |
| UC3239 | FIA | NG | FIA,FIB | NG | L/M | A/C |
| UC3521 | FIC | FIA,FIB | FIA,FIB | NG | L/M | A/C |
| UC3649 | FIA | FIA,FIB | FIA,FIB | P | L/M | A/C |
| UC3741 | NG | FIA,FIB | FIA,FIB | NG | L/M | A/C |
| UC4008 | FIC | NG | FIA,FIB | NG | L/M | A/C |
| UC4330 | FIB | NG | FIA,FIB | NG | L/M | A/C |
Discussion
E.coli is a common cause of community-acquired and health-care–acquired infections. Carbapenems are being increasingly used to treat infections due to multi drug resistant Enterobacteriaceae and sometimes empirically. This has got a major impact in the emergence of multi drug resistance which can be easily transmitted from one species to another by transferable elements such as plasmids. MIC values for imipenem, meropenem and ertapenem ranged from 8-64 µg/ml. Strains found to harbor both blaNDM-1 and blaOXA-48 showed higher MICs against carbapenems (64 µg/ml) as compared to MICs (8-16 µg/ml) showed by strains harboring blaNDM-1 only. Isolates were found to be susceptible to tigecycline and colistin as per MIC breakpoints. In this study, 45 (100%) blaNDM-1 positive E. coli isolates showed positive results from the modified Hodge test while finding from Castanheira M et al., reported the occurrence of weakly positive results for the modified Hodge test in the detection of NDM-1 producing Enterobacteriaceae [11].There was a 100% correlation with positive DDST, CDST and MBL (IP/IPI) E-test method with the presence of NDM-1in these clinical isolates as detected by PCR. The overall co-presence of blaOXA-48 and blaNDM-1 among E. coli in our study was was found to be (25/300) 8.3%. Among ESBL blaCTX-M-15 was the commonest genotype 38/45 (84%) followed by blaTEM 32/45(71%) blaSHV 28/45 (62%) and blaOXA 19/45(42%) either alone or in combination in the blaNDM-1 producing E.coli. Previous studies from India had reported the presence of TEM-1, CTX-M-15, SHV-1, SHV-12, DHA and CMY-2 and in the NDM-1 producing Enterobacteriaceae [12,13].While findings from other studies from abroad had showed the presence of blaCTX-M-15, blaTEM-1, blaSHV-28, blaSHV-11, and blaCMY-6 in the blaNDM-1 possessing Enterobacteriaceae [14,15].Though, the strain remains sensitive for tigecycline in vitro but it is not recommended for use in UTI infections. Colistin is the main stay of therapy.
Conclusion
Both blaNDM-1 and blaOXA-48 resulted in higher MICs against carbapenems (64 µg/ml) than presence of blaNDM-1 alone (>8-32µg/ml). This must be extremely worrisome, as dissemination of plasmids carrying resistant determinant genes from one species to another makes organism refractory to the common antibiotics used in clinical practice. Here we report the co-presence of NDM-1 with OXA-48 producing E.coli in urine culture from a tertiary care centre in central India. Early detection of these resistant determinant genes by molecular methods is essential in limiting the spread of infection due to these organisms.
[1]. DM Livermore, N Woodford, The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and AcinetobacterTrendsMicrobiol. 2006 14:413-420. [Google Scholar]
[2]. JG Collee, RS Miles, B Wan, Tests for the identification of bacteria. In: Collee JG, Fraser AG, Marmion BP, Simmons A editor. Mackie and Mc Cartney Practical Medical Microbiology. 1996 14th EditionEdinburghChurchill Livingstone:131-150. [Google Scholar]
[3]. Clinical and Laboratory Standards Institute.Performance Standards for Antimicrobial Susceptibility Testing: twenty second Informational Supplement M100-S22.. 2012 Wayne, PA, USACLSI [Google Scholar]
[4]. European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters (Version 2, January 1, 2012).http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_2.0_120221.pdf [Google Scholar]
[5]. K Lee, YS Lim, D Yong, JH Yum, Y Chong, Evaluation of the Hodge Test and the Imipenem-EDTA Double-Disk Synergy Test for differentiating metallo –betaEvaluation of the Hodge Test and the Imipenem-EDTA Double-Disk Synergy Test for differentiating metallo –betalactamase producing Isolates of Pseudomonas spp. And Acinetobacter spp. J Clin Microbiol. 2003 41:4623-6. [Google Scholar]
[6]. D Yong, K Lee, JH Yum, HB Shin, GM Rossolini, Y Chong, Imipenem EDTA disc method for differentiation of metallo beta lactamase producing clinical isolates of Pseudomonas spp. and Acinetobacter spp.J Clin Microbiol 2002 40:3798-801. [Google Scholar]
[7]. K Kumarasamy, MA Toleman, TR Walsh, J Bagaria, F Butt, R Balakrishnan, Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological studyLancet Infect Dis. 2010 10:597-602. [Google Scholar]
[8]. J Versalovic, T Koeuth, R Lupski J, Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomesNucleic Acids Res.. 1991 19:6823-31. [Google Scholar]
[9]. L Vogel, G Jones, S Triep, A Koek, L Dijkshoorn, RAPD typing of Klebsiella pneumoniae, Klebsiella oxytoca, Serratia marcescens andPseudomonas aeruginosa isolates using standardized reagents.Clin. Microbiol.Infect 1999 5:270-76. [Google Scholar]
[10]. A Carattoli, A Bertini, L Villa, V Falbo, KL Hopkins, EJ Threlfall, Identification of plasmids by PCR-based replicon typing methods.J. Microbiol 2005 63:219-28. [Google Scholar]
[11]. M Castanheira, LM Deshpande, D Mathai, JM Bell, RN Jones, RE Mendes, Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007.Antimicrob Agents Chemother 2011 55:1274-78. [Google Scholar]
[12]. C Lascols, M Hackel, SH Marshall, AM Hujer, S Bouchillon, R Badal, The increasing prevalence and the dissemination of NDM-1 metallo-β-lactamase in India: data from the SMART study (2009).J Antimicrob Chemother 2011 66:1992-97. [Google Scholar]
[13]. AU Khan, P Nordmann, NDM-1-producing Enterobacter cloacae and Klebsiella pneumoniae in diabetic foot ulcers in IndiaJ Med Microbiol 2012 61:454-56. [Google Scholar]
[14]. O Samuelsen, CM Thilesen, L Heggelund, AN Vada, A Kümmel, A Sundsfjord, Identification of NDM-1-producing Enterobacteriaceae in Norway.J Antimicrob Chemother. 2011 66:670-2. [Google Scholar]
[15]. L Poirel, G Revathi, S Bernabeu, P Nordmann, Detection of NDM- 1-producing Klebsiella pneumoniae in KenyaAntimicrob Agents Chemother 2011 55:934-36. [Google Scholar]