Background: Cystic Fibrosis (CF) is an autosomal recessive genetic disorder in white populations caused by mutation in a gene that encodes Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) protein. Since frequent respiratory tract infections are the major problem in patients with CF, obligation to identify the causative bacteria and determining their antibiotic resistance pattern is crucial. The purpose of this project was to detect Gram-negative bacteria (GNB) isolated from sputa of CF patients and to determine their antibiotic resistance pattern.
Materials and Methods: The sputum of 52 CF patients, treated as inpatients at hospitals in Tehran, was obtained between November 2011 and June 2012. Samples cultured in selective and non-selective media and GNB recognized by biochemical tests. Antimicrobial susceptibility testing to cephalosporins, aminoglycosides and carbapenems was performed by disk diffusion method and MICs of them were measured. For phenotypic detection of carbapenemase and ESBLs production, the Modified Hodge test, double disk synergy test and the combined disk methods were performed. Subsequently, the genes encoding the extended spectrum beta-lactamases (blaPER, blaCTX-M) and carbapenemases (blaIMP-1, blaGES, blaKPC, blaNDM, blaVIM-1, blaVIM-2, blaSPM, blaSIM) in Gram negative bacteria were targeted among the resistant isolates by using PCR. PFGE was used to determine any genetic relationship among the Pseudomonas aeruginosa isolated from these patients.
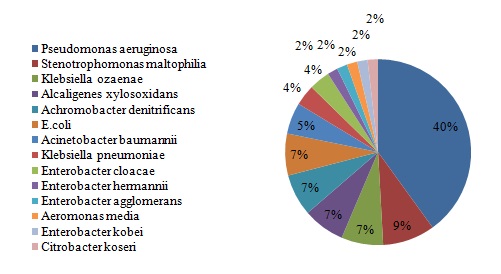
Results: Fifty five GNB were isolated from 52 sputum samples including Pseudomonas aeruginosa, Klebsiella ozaenae, Alcaligenes xylosoxidans, Achromobacter denitrificans, Klebsiella pneumonia and Stenotrophomonas maltophilia. The rates of resistance to different antibiotic were as follows: cefixime (%80), ceftriaxone (%43), ceftazidime (%45) and meropenem (%7). The prevalence of genes encoding the ESBLs and Carbapenemases among the the phenotypically positive strains were as follows: blaCTX-M (19), blaIMP-1 (2), blaVIM-1 (2) and blaVIM-2 (3) genes respectively. No other genes were detected. PFGE analysis revealed 8 genotypes. Six isolates had mutually 3 similar patterns.
Conclusion: This study showed the existence of important ESBLs and carbapenemases genes among the GNB isolated from patients with CF. Continuous surveillance of ESBLs and Carbapenemases, also identification of their types, in bacteria isolated from these patients have an important clinical impact, since, it can often provide valuable information for effective infection control measures and for the choice of appropriate antimicrobial therapy.
Introduction
CF is a severe autosomal recessive genetic disease that was first described in 1936 by the Swiss pathologist, Guido Fanconi, who reported the autopsy and clinical characteristics of three patients with bronchiectasis and pancreatic insufficiency [1]. In 1938, Dorothy Andersen published an autopsy study of 38 infants, described the findings as “cystic fibrosis of the pancreas” and recognized the syndrome as an inherited disease [2].
The abnormal salt transport in CF became clear during a heat wave in 1952, when children with CF were admitted to hospital with severe dehydration and salt loss [3]. The mechanism for the defect chloride transport in the sweat glands was demonstrated in 1983 [4], and in 1985 the gene was localized to the long arm of chromosome 7. This large gene named CF transmembrane conductance regulator (the CFTR gene), consists of 27 coding exons [5]. The person dies by progressive bronchiectasis and chronic respiratory insufficiency [6,7]. Failure of innate defense mechanisms and the lack of mucocilliary clearance in the airways stimulate primary and recurrent bacterial infections, blockage of airways, inflammation and chronic bacterial infections [8,9]. The bacterial species most commonly associated with respiratory tract infection in CF include common human pathogens such as Staphylococcus aureus and Haemophilus influenzae as well as several opportunistic pathogens, the most important of which is Pseudomonas aeruginosa [10]. With the improved survival, new emergent pathogens in the CF lung as Burkholderia cepacia complex (BCC), Stenotrophomonas Maltophilia (SM) and Alcaligenes Xylosoxidans (AX) have been detected in the last years [11]. It is possible to prevent or delay the onset of chronic infections in most patients with CF by eliminating cross-infections and by early aggressive antibiotic treatment of the positive sputum culture [12]. The most antibiotic used for these patients are β-lactams, aminoglycosides, andcorticosteroids. On the other hand microbial resistance against oxyimino-cephalosporins such as ceftriaxone and Ceftazidime (CAZ) and carbapenems is a growing problem in the treatment of infections. This is often caused by the production of Extended-Spectrum Betalactamases (ESBLs) and carbapenemases. ESBLs are frequently identified in Klebsiella pneumonia and Escherichia coli [13], but also in other species, such as Citrobacter spp., and Pseudomonas aeroginosa. The most abundant types are represented by SHV, TEM, and CTX-M [14].
The IMP- and VIM-type enzymes are two major groups of carbapenemases [15]. IMP-1 was the first identified acquired MBL [16] and has spread among Enterobacteriaceae, Pseudomonas aeruginosa, and other nonfastidious Gram-negative nonfermenters in Japan [17–20].
VIM-1 was identified from a clinical isolate of P. aeruginosa in Italy [21], and outbreaks of the VIM-1-producing P. aeruginosa isolates have been recognized in Greece [22] as well as Italy [23]. VIM-2 was firstly identified from a clinical isolate of P. aeruginosa in France [24]. In this study, we analyzed the pathogens associated with respiratory tract infection in CF patients and investigated the production of ESBLs and carbapenemases. We also used pulse field gel electrophoresis (PFGE) to investigate the possible genetic relationship among the Pseudomonas aeruginosa strains colonizing the respiratory tract of these patients.
Material and Methods
Patients and bacterial strains
Sputum of patients with CF treated as in patients at two hospitals in Tehran were collected from November 2011 to June 2012. Ethical clearance was obtained before the collection of samples. Clinical specimens obtained by throat swab or oropharyngeal suction, were cultured in selective and non selective media including (Blood, Chocolate, EMB, MacConkey) agar, Burkholderiacepacia Special Agar (BCSA) and Mueller Hinton agar. Isolates were identified based on the biochemical tests and/or the API20E system (France-BioMerieux).
Susceptibility tests
Susceptibility to antibiotics were measured by Kirby Bauer disk diffusion method. The MICs of resistant isolates to cefotaxime, ceftriaxone, imipenem and kanamycin were determined using broth microdilution method. Both tests were performed and interpreted according to the CLSI guidelines (The Clinical and Laboratory Standards Institute). The tested antimicrobial agents (MAST, Co., UK) were Ceftazidime (CAZ), Cefotaxime (CTX), Cefixime (CFM), Cefepime (CFM), Gentamicin (GM), Amikacin (AK), PolymyxinB (PB), Piperacillin/tazobactam (PTZ), Ciprofloxacin (CIP), Ceftriaxone (CRO), Meropenem (MEM), Imipenem (IMP), Ertapenem (ERT). Pseudomonas aeroginosa ATCC27853 was used as positive control.
ESBL activity
ESBL activity in isolates showing resistance to cefotaxime was undertaken by disk synergy testing of CTX in the presence and absence of clavulanic acid which were placed on a plate of Mueller Hinton agar inoculated with suspension (turbidity of 0.5 MacFarland) of isolates. E. coli (ATCC25922) was used as positive control reference strain. A positive test result was defined as a ≥ 5 mm difference in the zone diameter between two disks [25].
Screening of Carbapenemase producers
Carbapenem resistant isolates were subjected to a screening test for MBL production using EDTA-disk synergy test and modified Hodge test according to Lee et al., [26] instructions.
DNA extraction: DNA templates for polymerase chain reaction PCR was extracted by suspending 4-5 colonies of an overnight growth of isolate on Mueller Hinton agar, in 500μl of double distilled water. The suspension was boiled at 100°C for 10 minutes and frozen for 5 minutes. Then, it was centrifuged at 19000 rpm for 5 minutes. An aliquot in 1μl of the supernatant was used as DNA template for PCR [27].
PCR procedure:
Isolates included in this study were screened by PCR for the following ESBLs and carbapenemases encoding genes: ESBLs (blaPER, blaCTX-M) and Carbapenemases (blaIMP-1, blaGES, blaKPC, blaNDM, blaVIM-1, blaVIM-2, blaSPM, blaSIM). K. pneumonia 7881 containing blaCTX-M, P. aeruginosa KOAS containing blaPER gene, A. baumannii AC54/97 producing blaIMP gene, P. aeruginosaPO510 producing blaVIM-1, P. aeruginosa COL-1 producing blaVIM-2 and P. aeruginosa16 producing blaSPM-1 (Kindly provided by Patrice Nordmann)strains were used as positive controls. The sequences of primers are shown in [Table/Fig-1].
Primers used for PCR amplification of different genes encoding ESBLs and carabapenemases
| Primer | Gene | Sequence (5’-3’) | Size (bp) | References |
|---|
| CTXM-F | blaCTX-M | CGCTTTGCGATGTGCAG | 550 | [28] |
| CTXM-R | ACCGCGATATCGTTGGT |
| PER-F | blaPER | AATTTGGGCTTAGGGCAGAA | 925 | [29] |
| PER-R | ATGAATGTCATTATAAAAGC |
| KPC-F | blaKPC | CTTGCTGCCGCTGTGCTG | 489 | [30] |
| KPC-R | GCAGGTTCCGGTTTTGTCTC |
| GES-F | blaGES | ATGCGCTTCATTCACGCAC | 840 | [31] |
| GES-R | CTATTTGTCCGTGCTCAGG |
| NDM-F | blaNDM | ACCGCCTGGACCGATGACCA | 263 | [32] |
| NDM-R | GCCAAAGTTGGGCGCGGTTG |
| VIM1-F | blaVIM-1 | AGTGGTGAGTATCCGACA | 261 | [33] |
| VIM1-R | ATGAAAGTGCGTGGAGAC |
| VIM2-F | blaVIM-2 | ATGTTCAAACTTTTGAGTAAG | 801 | [33] |
| VIM2-R | CTACTCAACGACTGAGCG |
| IMP1-F | blaIMP-1 | ACCGCAGCAGAGTCTTTGCC | 587 | [33] |
| IMP1-R | ACAACCAGTTTTGCCTTACC |
| SPM-F | blaSPM | GCGTTTTGTTTGTTGCTC | 786 | [33] |
| SPM-R | TTGGGGATGTGAGACTAC |
| SIM-F | blaSIM | TACAAGGGATTCGGCATCG | 571 | [34] |
| SIM-R | TAATGGCCTGTTCCCATGTG |
| OXA48 | blaOXA-48 | TTGGTGGCATCGATTATCGG | 743 | [35] |
| OXA48 | GAGCACTTCTTTTGTGATGGC |
To identify the size of genes, PCR products were run on %1 agarose gel and visualized by gel documentation.
Genotyping
DNA typing was performed by PFGE according to the protocol described by Nikbin et al., [36]. PFGE has been widely used to type various microorganisms in both outbreak and population based studies. The percentage of relatedness were calculated by usage of the Dice coefficient. DNA patterns were aslo analyzed virtually as instructed by Tenover et al [37]. Accordingly, strains with up to three band differences were considered closely related, strains with four to six band differences were considered possibly related, and strains with greater than six band differences were considered unrelated.
Results
Study population
The study population consisted of 52 CF patients from two Paediatric hospitals in Tehran. Patients ranged in ages from 1.5 months to 16 years. The male: female ratio was 2:1.
Sample processing
All throat swabs, oropharyngeal suctions and sputa were cultured. A total of 55 GNB were isolated from 52 samples. Six samples contained just Gram-positive pathogens. Also 1 fungus was isolated. No organisms were isolated from 5 samples.
Fifty five isolates belonging to different species of GNB were isolated [Table/Fig-2].
Antimicrobial susceptibility testing
The results of susceptibility on 55 isolates of GNB are shown in [Table/Fig-3].
The number of resistant bacteria to different antibiotics
| Antibiotic | Number of resistant isolates (%) |
|---|
| Cefotaxime | 26(47.3%) |
| Ceftazidime | 25(45.5%) |
| Ceftriaxone | 24(43.6%) |
| Cefexime | 44(80%) |
| Imipenem | 7(12.7%) |
| Meropenem | 11(20%) |
| Gentamicin | 14(24.5%) |
| Ciprofloxacin | 10(18%) |
| Piperacillin/Tazobactam | 11(20%) |
Anti-biogram showed that 89.09% of isolates were resistant to at least one of the third generation cephalosporins.
MIC tests showed that 40% [22] of isolates were resistant to cefotaxime. Of these, 16 (72.72%) were positive in combined disk tests for ESBLs detection. Eight isolates (14.51%) were resistant to meropenem. The modified Hodge test was positive in 3 isolates. The EDTA disk synergy was also positive in 4 of 11 imipenem resistant isolates.
PCR: PCR demonstrated that 19 of isolates contained blaCTX-M.Of 17 carbapenem resistant isolates, blaIMP-1were detected in 2, blaVIM-1 in 2 and blaVIM-2 in 3 respectively. No other genes were detected.The number of isolates within every species containing blaCTX-M, blaIMP-1, blaVIM-1, blaVIM-2 and blaPE Rare shown in [Table/Fig-4].
Prevalence of some of ESBL and Carbapenemase genes in isolates.
| Isolates | blaCTX-M | blaIMP-1 | blaVIM-1 | blaVIM-2 |
|---|
| P. aeruginosa | 5 | - | - | - |
| K.ozaenae | 2 | - | - | - |
| A. xylosoxidans | 2 | 2 | 2 | 3 |
| A. denitrificans | 1 | - | - | - |
| E.coli | 4 | - | - | - |
| S. maltophilia | - | - | - | - |
| A. baumannii | - | - | - | - |
| K. pneumonia | - | - | - | - |
| E. cloacae | 1 | - | - | - |
| E. hermannii | 1 | - | - | - |
| E. agglomerans | 1 | - | - | - |
| Aeromonas media | - | - | - | - |
| E. kobei | 1 | - | - | - |
| C. koseri | 1 | - | - | - |
PFGE: The PFGE patterns obtained from 11 strains of P. aeruginosa are shown in [Table/Fig-5]. It was used to assess the clonality of them. The PFGE analysis revealed 8 different clusters. Of these isolates 6 were mutually similar. They were collected from one hospital. 3 clusters from 3 isolates obtained from another hospital were completely different. No similarity were seen between isolates of two hospitals.
Dendrogram of PFGE results for P. aeruginosa isolates. It revealed 8 different clusters
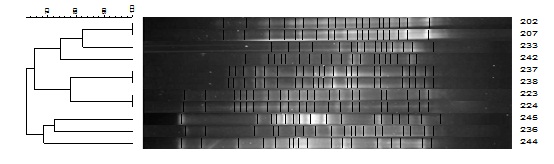
Of these isolates 6 were mutually similar. They were collected from one hospital. 3 clusters from 3 isolates obtained from another hospital were completely different. No similarity were seen between isolates of two hospitals.
Discussion
CF is the most common life-shortening autosomal recessive disease in the white population and afflicts about 60,000 patients worldwide, approximately 30,000 of whom are cared for in the United States [11]. The abnormal characteristic of this disease is the movement of water and ions through the epithelial cells that leads to formation of a dense mucosa and decrease in mucosal clearance in the lungs [38].
The microbiology of CF pulmonary infection has changed over the past 5 decades, as modern therapies enable patients with CF to live longer. Unusual pathogens and highly antibiotic-resistant organisms are increasingly recovered from patients with more advanced disease.
One of the major aspects of this study is reporting the broad range of bacteria identified in sputum and throat swab cultures of 52 CF patients. We isolated P. aeruginosa, K. zaenae, A. xylosoxidans, A. denitrificans, K. pneumoniae, S. maltophilia, E. hermannii, E. agglomerans, E. cloacae, E. kobei and C. koseri. As reported by Paixão et al., in 2010, P. aeruginosa as is the most frequent pathogen in CF patients [39]. We obtained the same result. Studies from Iran concerning the infective microorganisms among patients with CF is limited. In 2006 and 2010 Eftekhar et al., [40] and Khanbabaei et al., [41], detected P. aeruginosa as the most common agent and they reported that 85.7% of microorganisms were susceptible to ceftazidime. In another research in Iran which was done by Forozeshfard et al., [42], 72% of P. aeruginosa isolates from sputa of CF patients were susceptible to ceftazidime, and none of them showed resistance to imipenem. While in this study 44.45% of pathogens were resistant to ceftazidime. The reason might be extra administration and usage of ceftazidime and/or obtaining resistant genes which was widelyspread in the hospital by pathogens causing infection in CF patients.Fortunately in all studies including ours, imipenem was the most effective antibiotic. This may be due to limited administration of this drug tothese patients.Among a variety of drug-resistance traits, ESBL-producing GNB with resistance to newer cephalosporins have been posing a significant challenge in clinical practice. CTX M-1 had been observed in P. aeruginosa and S. Maltophilia isolated from patients with CF in Greece [43]. Multidrug-resistant P. aeruginosa isolate co-expressing extended-spectrum β-lactamase PER-1 and metallo-β-lactamase VIM-2 had been recovered from a 2-year-old child suffering from pneumonia in an underlying context of CF in Turkey [44]. A VIM-2 enzyme was detected in P. aeruginosa isolates in CF patents in Portugal [45].
The genes encoding carbapene mases and ESBLs were detected among the isolates in this study too. Carbapenemases represent the most versatile family of β-lactamases, with a breadth of spectrum unrivaled by other β-lactam-hydrolyzing enzymes. Until the early 1990s, all carbapenemases were described as species-specific, chromosomally encoded β-lactamases, each with a well-defined set of characteristics.
In a study in Iran, strains of P. aeruginosa isolated from CF patients were checked for production of MBLs using PCR targeting blaVIMand none of clinical isolates was positive for it [42]. Among MBL genes, VIM-type had been detected in P. aeruginosa in Iran [46], NDM-1 in K. pneumonia [47] and SPM-1, GES-1, OXA-23,OXA51 in A. baumanii [48]. In the current study, we identified some of the genes encoding Carbapenemases and ESBLs among the isolates and these were blaPER, blaIMP-1, blaVIM-1blaVIM-2and blaCTX-M. With the incidence of 34.54% (n=19), blaCTX-Mwas the most prevalent gene. Despite the prevalence of genes encoding carbapenemases and ESBLs in Iran, this is the first description of them which were isolated from CF patients. The clonal relationship between P. aeruginosa isolates was studied by PFGE and while 6 isolates were mutually related, a close relationships among 7 isolates was observed. Three CTX-M producing isolates showed closely related patterns.They belonged to the same hospital. Other isolates (n=4) were genetically distinct.
In conclusion, as the transmission of isolates in CF patients is not well specified, therefore it is important to separate patients, allocate a special center for them and design infection control policies. Also we suggest that careful supervision of the prevalence of antibiotic resistance in these patients should be established.
[1]. Fanconi G, Wehlinger E, Knauer C, Das Cocliakie-Syndrom bei bronchiectasienWien Med Wochenschr 1936 86:753-56. [Google Scholar]
[2]. Andersen DH, Cystic fibrosis of the pancreas and its relation to celiac disease: A clinical and pathological studyAm J Dis Child 1938 56:344-49. [Google Scholar]
[3]. Di Sant’, Agnese PA, Darling RC, Perera GA, Shea E, Abnormal electrolyte composition in sweat in cystic fibrosis of the pancreas, its clinical significance and relationship to the diseasePaediatrics 1953 12:549-63. [Google Scholar]
[4]. Quinton PM, Chloride impermeability in cystic fibrosisNature 1983 301:421-22. [Google Scholar]
[5]. Amaral MD, Processing of CFTR: traversing the cellular maze—how much CFTR needs to go through to avoid cystic fibrosis?Paediatr Pulmonol 2005 39:479-91. [Google Scholar]
[6]. Chaparro C, Maurer J, Gutierrez C, Krajden M, Chan C, Winton T, Infection with Burkholderiacepacia in cystic fibrosis: Outcome following lung transplantationAm J Respir Crit Care Med 2001 163:43-48. [Google Scholar]
[7]. Goldman L, Bennett JC, CECIL: Textbook of Medicine 2001 Rio de JaneiroGuanabara Koogan [Google Scholar]
[8]. Accurso FJ, Update in cystic fibrosis 2005Am J Respir Crit Care Med 2006 173:944-47. [Google Scholar]
[9]. Boucher RC, New concepts of the pathogenesis of cystic fibrosis lung diseaseEur Respir J 2004 23(1):146-58. [Google Scholar]
[10]. Lipoma JL, The changing microbial epidemiology in cystic fibrosisClin Microbiol Rev 2010 23(2):299-323. [Google Scholar]
[11]. Saiman L, Siegel J, Infection control in cystic fibrosisClin Microbiol Rev 2004 17(1):57-71. [Google Scholar]
[12]. Saiman L, MacDonald N, Burns JL, Hoiby N, Speert DP, Weber D, Infection control in cystic fibrosis: pratical recommendations for the hospital, clinic, and social settingAm J Infect Control 2000 28:381-85. [Google Scholar]
[13]. Gniadkowski M, Evolution and epidemiology of extended-spectrum β-lactamases (ESBLs) and ESBL-producing microorganismsClin Microbiol Infect 2001 7:597-608. [Google Scholar]
[14]. Gniadkowski M, Schneider I, Jungwirth R, Hryniewicz W, Bauernfeind A, Ceftazidime resistant Enterobacteriaceaeisolates from three Polish hospitals: identification of three novel TEM- and SHV-5-type extended-spectrum β-lactamasesAntimicrob Agents Chemother 1998 42:514-20. [Google Scholar]
[15]. Livermore DM, N Woodford, Carbapenemases: a problem in waiting?Curr. Opin. Microbiol 2000 3:489-95. [Google Scholar]
[16]. Osano E, Arakawa Y, Wacharotayankun R, Ohta M, Horii T, Ito H, Molecular characterization of an enterobacterialmetallo-β-lactamase found in a clinical isolates of Serratiamarcescensthat shows imipenem resistanceAntimicrob. Agents Chemother 1994 38:71-78. [Google Scholar]
[17]. Hirakata Y, Lzumikawa K, Yamaguchi T, Takemura H, Tanaka H, Yoshida R, Rapid detection and evaluation of clinical characteristics of emerging multiple-drug-resistant gram-negative rods carrying the metallo-β-lactamase gene blaIMPAntimicrob Agents Chemother 1998 42:2006-2011. [Google Scholar]
[18]. Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M, Plasmid-mediated dissemination of the metallo-13-lactamase gene blaIMP among clinical isolated strains of SerratiamarcescensAntimicrob Agents Chemother 1995 39:824-29. [Google Scholar]
[19]. Senda K, Arakawa Y, Nakashima K, Ito H, Ichiyama S, Shimokata K, Multifocal outbreaks of metallo-β-lactamase-producingPseudomonasaeruginosaresistant to broad-spectrum-β-lactams, including carbapenemsAntimicrob. Agents Chemother 1996 40:349-53. [Google Scholar]
[20]. Senda K, Arakawa Y, Ichiyama S, Nakashima K, Ito H, Ohsuka S, PCR detection of metallo-β- lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum β-lactamsJ Clin Microbiol 1996 34:2909-13. [Google Scholar]
[21]. Lauretti L, Riccio ML, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Cloning and characterization of blaVIM, a newintegron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosaclinical isolateAntimicrob Agents Chemother 1999 43:1584-90. [Google Scholar]
[22]. Tsakris A, Pournaras S, Woodford N, Palepou MFI, Babini GS, Douboyas J, Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in GreeceJ. Clin. Microbiol 2000 38:1290-92. [Google Scholar]
[23]. Cornaglia G, Mazzariol A, Lauretti L, Rossolini GM, Fontana R, Hospital outbreak of carbapenem-resistant Pseudomonas aeruginosaproducing VIM-1, a novel transferable metallo-β-lactamaseClin. Infect. Dis 2000 31:1119-25. [Google Scholar]
[24]. Poirel L, I Le Thomas, Naas T, Karim A, Nordmann P, Biochemical sequence analyses of GES-1, a novel class a extended-spectrum β-lactamase, and the Class 1 Integron In52 from Klebsiella pneumoniaAntimicrob Agents Chemother 2000 44(3):622-32. [Google Scholar]
[25]. NCCLSZone diameter interpretive standardsNCCLS global information supplement 2001 21:40-71. [Google Scholar]
[26]. Lee K, Lim YS, Yong D, Yum JH, Chong Y, Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase producing isolates of Pseudomonas spp. and Acinetobacter spp.J Clin Microbiol 2003 41:4623-29. [Google Scholar]
[27]. Barguigua A, El Otmani F, Talmi M, Boutjilat F, Haouzane K, Timinouni M, Characterization of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumonia isolates from the community in Morocco 2011 60:1344-52. [Google Scholar]
[28]. Ahmed MA, Nakano H, Shimamoto T, The first characterization of extended-spectrum β-lactamase-producing Salmonella in JapanJ. Antimicrob. Chemother 2004 54(1):283-84. [Google Scholar]
[29]. Claeys G, Verschraegen G, de Baere T, Vaneechoutte M, PER-1 β- lactamase-producing Pseudomonas aeruginosa in an intensive care unitJ Antimicrob Chemother 2000 45(6):924-25. [Google Scholar]
[30]. Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typingJ Clin Microbiol 1995 [Google Scholar]
[31]. Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid-and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in FranceAntimicrob Agents Chemother 2000 44:891-97. [Google Scholar]
[32]. Zarfel G, Hoenigl M, Leitner M, Salzer H, Feierl G, Masoud L, Emergence of New Delhi Metallo-β-LactamaseEmerg Infect Dis 2011 17(1):129-30. [Google Scholar]
[33]. Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, PCR typing of genetic determinants for metallo-β-lactamases and integrases carried by gram-negative bacteria isolated in Japan, with focus on the Class 3 IntegronJ Clin Microbiol 2003 41(12):5407-13. [Google Scholar]
[34]. Lee K, Yum JH, Yong D, Lee HM, Chong Y, Novel acquired metallo-β-lactamase gene, blaSIM-1, in a Class 1 Integron from Acinetobacter baumannii clinical isolates from KoreaJ Antimicrob Chemoter 2005 49(11):4485-91. [Google Scholar]
[35]. Aktas Z, Satana D, Kayacan C, Ozbek B, Carbapenem resistance in Turkey: Repeat report on OXA-48 in Klebsiella pneumoniae and first report on IMP-1 beta-lactamase in Escherichia coliAfrican Journal of Microbiology Research 2012 6(17):3874-78. [Google Scholar]
[36]. Nikbin VS, Abdi-Ali A, Feizabadi MM, Gharavi S, Pulsed field gel electrophoresis & plasmid profile of Pseudomonas aeruginosa at two hospitals in Tehran, IranIndian J Med Res 2007 146:15 [Google Scholar]
[37]. Tenover F, Kalsi R, Williams P, Carey R, Stocker S, Lonsway D, Carbapenem Resistance in Klebsiellapneumoniae not detected by automated susceptibility testingEmerg Infect Dis 2006 12(8):1209-13. [Google Scholar]
[38]. Govan JR, Deretic V, Microbial pathogenesis in cystic fibrosis: Mucoid Pseudomonas aeruginosa and BurkholderiacepaciaMicrobiol Rev 1996 60:539-74. [Google Scholar]
[39]. Paixao VA, Barros TF, Mota CM, Moreira TF, Santana MA, Reis JN, Prevalence and antimicrobial susceptibility of respiratory pathogens in patients with cystic fibrosisBraz J Infect Dis 2010 14(4):406-9. [Google Scholar]
[40]. Eftekhar F, Rostamizadeh F, Khodadad A, Henry D, Speert DP, Isolation and genetic fingerprinting of Pseudomonas aeruginosa from Iranian patients with cystic fibrosis using RAPD-PCRIranian Journal of Biotechnology 2003 1(2) [Google Scholar]
[41]. Khanbabaee Gh, Akbarizadeh M, Sayyari A, Ashayeri-Panah M, Abdollahgrji F, A survey on pulmonarypathogens and their antibiotic susceptibility among cystic fibrosis patientsBraz J Infect Dis 2012 16(2) [Google Scholar]
[42]. Forozesh Fard M, Irajian G, MoslehiTakantape Z, Fazeli H, Salehi M, Rezania S, Drug resistance pattern of Pseudomonas aeruginosa strains isolated from Cystic fibrosis patients at Isfahan AL Zahra hospital, Iran 2012 4:97 [Google Scholar]
[43]. Rao S. CTX-M β-lactamases. Department of Microbiology, School of Medicine, University of Zagreb Clinical Department of Clinical and Molecular Microbiology. 2012; 33: 2233–39 [Google Scholar]
[44]. Yakupogullari Y, Poirel L, Bernabeu S, Kizirgil A, Nordman P, Multidrug-resistant Pseudomonas aeruginosa isolateco-expressing extended-spectrum β-lactamase PER-1 and metallo-β-lactamase VIM-2 from TurkeyJournal of Antimicrobial Chemotherapy 2007 [Google Scholar]
[45]. Cardoso O, Alves AF, Leitão R, Metallo-beta-lactamase VIM-2 in Pseudomonas aeruginosa isolates from a cystic fibrosis patientInt J Antimicrob Agents 2008 31(4):375-9. [Google Scholar]
[46]. Shahcherghai F, Nikbin VS, Feizabadi MM, Identification and genetic characterization of metallobeta-β lactamase-producing strains of Pseudomonas aeruginosain Tehran, IranNew Microbiol 2010 33:243-48. [Google Scholar]
[47]. Shahcheraghi F, Abbasalipour M, Feizabadi MM, Ebrahimipour GH, Akbari N, Isolation and genetic characterization of metallo-β-lactamase and carbapenamase producing strains of Acinetobacterbaumannii from patients at Tehran hospitalsIran J Microbiol 2011 3(2):68-74. [Google Scholar]
[48]. Shahcheraghi F, Nobari S, Rahmati Ghezelgeh F, Nasiri S, First report of New Delhi metallo-beta-lactamase-1-producing Klebsiellapneumoniae in IranMicrobial Drug Resistsance, Mary Ann Liebert, Inc 2013 19(1) [Google Scholar]