Extended spectrum β-lactamase (ESBL) isolates were first detected in Western Europe in the mid-1980s. Since then, their incidence has been increasing steadily. ESBLs are able to hydrolyze 3 and 4 generation cephalosporins and monobactams. ESBL producing strains are inhibited by β-lactamase inhibitors (clavulanic acid, sulbactam and tazobactam) [1,2]. A large number of outbreaks of infections due to ESBL producing organisms have been described on every continent of the globe. In some hospitals, initial outbreaks of infections have been supplanted by endemicity of the ESBL producing organisms. This may lead to increased patient mortality when antibiotics inactive against ESBL producers are used. Therefore, control of the initial outbreak of ESBL producing organisms in a hospital or specialized unit of a hospital is of critical importance [3–5].
ESBLs are a group of enzymes encoded by genes described predominantly on plasmid that are common among Enterobacteriaceae [6]. Although most ESBLs are mutants of temoneira (TEM) and sulfhydryl variable (SHV) enzymes, the cefotaximase (CTX-M) type-lactamases which have become important, originated from β-lactamases found in environmental species of the genus Kluyvera, and this enzyme hydrolyzes cefotaxime and cefriaxone but is weakly active against ceftazidime [7,8]. At present, there are more than 300 different ESBL variants, and these have been clustered into nine different structural and evolutionary families based on amino acid sequence. TEM and sulphydryl variable SHV were the major types. However, CTX-M type is more common in some countries [9].
Determination of TEM and SHV genes by molecular techniques in ESBL producing bacteria and their pattern of antimicrobial resistance can supply useful data about their epidemiology and risk factors associated with these infections [10]. The aim of this study was to isolate and identify ESBL producing bacteria and then further molecular characterization of the types of extended spectrum β-lactamases (ESBL) produced by E. coli, and Klebsiella spp.
Material and Methods
Different clinical specimens such as blood, urine, respiratory, pus, sterile body fluids including ascitic fluid, CSF, gastric aspirate, synovial fluid, peritoneal fluid, and stool samples were collected from different clinical specialties of a NIMS hospital, in rural area of Jaipur, Rajasthan, India from February, 2011 to March, 2012. 722 Gram negative bacilli (GNB) were isolated. Isolates showing reduced zone of inhibition to third generation cephalosporins i.e. ceftazidime (30μg), cefotaxime (30μg) and to fourth generation cephalosporins, cefepime (30μg) were screened for ESBL production.
ESBL detection: ESBL producing isolates were characterized phenotypically for ESBL production using double disc synergy test (DDST) as recommended by the Clinical Laboratory Standards Institute (CLSI) [11]. The test was done by using both cefotaxime (30μg) and ceftazidime (30μg) alone and in combination with clavulanic acid. A > 5 mm increase in zone diameter for either antimicrobial agent tested in combination with clavulanic acid versus its zone when tested alone was taken as positive result for ESBL production [Table/Fig-1].
K. pneumoniae ATCC 700603 (positive control) and E. coli ATCC 25922 (negative control) were used for quality control for ESBL tests.
Molecular Characterization of ESBL Producing Esherichia coli and Klebsiella spp.
Twenty ESBL producing isolates each of E. coli and K. pneumonia were selected for detection of β-lactamase encoding genes of the family TEM, SHV and cefotaximase CTX-M.
Plasmid DNA was isolated from bacterial cells by using PureSol TM Plasmid Isolation Kit (GeNeiTM Cat No.: 612116900021730) by using manufacturer instructions.
For PCR amplifications, master mix was prepared containing 200 mM of dNTPs (GeNeiTM Cat No: FC10L), 0.4 mM of each primer, 2.5 U of Taq polymerase (GeNeiTM Cat No: MME5J) in 1x PCR buffer. 500 pg of DNA was added and final volume of mixture was made up to 50 μl. Primers were custom designed for the study.
1. TEM-1 beta-lactamase
F. P: 5′-GAGACAATAACCCTGGTAAAT-3′
R. P: 5′ - AGAAGTAAGTTGGCAGCAGTG - 3′
2. CTX-M beta-lactamase
F P: 5-GAAGGTCATCAAGAAGGTGCG-3′
R P: 5′-GCATTGCCACGCTTTTCATAG-3′
3. SHV beta-lactamase
F. P: 5′-GTCAGCGAAAAACACCTTGCC-3′
R. P: 5′- GTCTTATCGGCGATAAACCAG - 3′
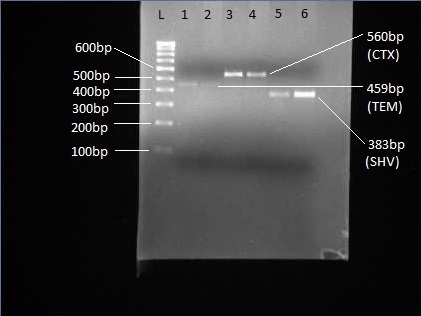
Amplification was performed in a Gradient MyCycler (Bio-Rad) with cycling parameters as mentioned in [Table/Fig-2]. Agarose Gel Electrophoresis was done at 50 volt for 2.5 hour. Gel was visualized on UV platform in Gel Documentation system XR+ (Bio-Rad, USA) using quantity one software [Table/Fig-3].
Cycling Parameters for Amplification
| Initial Denaturation | 95°C | 5 min | |
|---|
| Denaturation | 95°C | 30 sec | 35 cycles |
| Annealing | 550C for TEM, 600C for SHV and CTX | 30 sec |
| Elongation | 72°C | 2 min |
| Final elongation | 72°C | 10 min | |
PCR products of blaTEM ( Lines 1-2), blaCTX-M (Lines 3, 4) and blaSHV (Lines 5, 6 ) and L: 100 bp DNA ladder
Results
A total of 722 GNB were isolated during the study from different clinical samples [Table/Fig-4]. Among 722 GNB isolated 379 (52.49%) were ESBL producing while in 343 (47.51%) strains ESBL production was not found. ESBL producing GNB were isolated from various clinical samples urine (57.2%), blood (31.07%), pus (48.03%), respiratory tract (63.83%), body fluid (52.17%) and stool samples (59.29%) [Table/Fig-5]. Among various GNB isolated highest ESBL production was observed in Klebsiella spp. (67.04%) followed by Escherichia coli (56.92%), Proteus spp. (46%), Pseudomonas spp. (41.89%), Citrobacter freundii (27.59%), Salmonella typhi (26.31%), Acinetobacter spp (11.11%) and Salmonella paratyphi A (5.56%) [Table/Fig-5]. Among 379 ESBL producing 64 (16.89%) ESBL producers were isolated from OPD, 245 (64.64%) were isolated from wards and 70 (18.47%) ESBL producers were isolated from ICU.
Prevalence of Gram negative bacterial isolates from clinical samples
| Organism | Urine | Blood | Pus | Respiratory Isolates | Body Fluid | Stool | Total No. |
|---|
| Escherichia coli | 141 | 30 | 46 | 30 | 10 | 68 | 325 |
| Klebsiella sp. | 40 | 17 | 34 | 48 | 7 | 33 | 179 |
| Pseudomonas sp. | 11 | - | 42 | 14 | 6 | 1 | 74 |
| Proteus sp | 31 | - | 13 | - | - | 6 | 50 |
| Citrobacter fruendi | 11 | - | 12 | - | - | 6 | 29 |
| Acinetobacter sp | 2 | - | 5 | 2 | - | - | 9 |
| Salmonella Typhi | - | 38 | - | - | - | - | 38 |
| Salmonella paratyphi A | - | 18 | - | - | - | - | 18 |
| Total | 236 | 103 | 152 | 94 | 23 | 114 | 722 |
Prevalence of ESBL production among bacterial isolates from clinical samples
| Site → | Urine | Blood | Pus | Respiratory Isolates | Body Fluid | Stool | Total |
|---|
| Organism ↓ | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % |
| Escherichia coli | 82 | 58.16 | 13 | 43.33 | 28 | 60.87 | 21 | 70.00 | 6 | 60 | 35 | 51.47 | 185 | 56.92 |
| Klebsiella spp. | 27 | 67.50 | 8 | 47.06 | 22 | 64.71 | 33 | 68.75 | 3 | 42.86 | 27 | 81.82 | 120 | 67.04 |
| Pseudomonas spp. | 6 | 54.55 | - | - | 16 | 38.10 | 6 | 42.86 | 3 | 50 | - | - | 31 | 41.89 |
| Proteus spp. | 19 | 61.29 | - | - | 3 | 23.08 | - | - | - | - | 1 | 16.67 | 23 | 46.00 |
| Citrobacter fruendi | 1 | 9.09 | - | - | 3 | 25.00 | - | - | - | - | 4 | 66.67 | 8 | 27.59 |
| Acinetobacter spp. | - | 0.00 | - | - | 1 | 20.00 | 0 | 0.00 | - | - | - | - | 1 | 11.11 |
| Salmonella typhi | - | - | 10 | 26.31 | - | - | - | - | - | - | - | - | 10 | 26.31 |
| Salmonella paratyphi A | - | - | 1 | 5.56 | - | - | - | - | - | - | - | - | 1 | 5.56 |
| Total | 135 | 57.20 | 32 | 31.07 | 73 | 48.03 | 60 | 63.83 | 12 | 52.17 | 67 | 59.29 | 379 | 52.49 |
Antibiotic susceptibility pattern of ESBL isolates is shown in [Table/Fig-6]. All the isolates were sensitive to imipenam. Among β-Lactam/ β-Lactam inhibitor drugs highest sensitivity of was observed with cephoparazone/sulbactum in case of E. coli, Klebsiella spp. and Proteus spp, while in case of Citrobacter spp. and Pseudomonas spp. highest sensitivity was observed with Piperacillin/Tazobactum. Among aminoglycosides highest sensitivity was observed with netillin followed by amikacin and gentamycin in all isolates. Among quinolones sensitivity to ofloxacin is higher than ciprofloxacin. However, among enterobacteriaceae isolates more than 60% resistance was observed against quinolones.
Sensitivity pattern of ESBL isolates to various antibiotics. *Only one isolate
| Antibiotic → Organism Isolated ↓ | Cefepime | Amikacin | Gentamycin | Netillin | Ciprofloxacin | Ofloxacin | Amoxyclav | Cephoparazone/Sulbactum | Piperacillin/Tazobactum | Imipenem |
|---|
| Escherichia coli | 13.51 | 78.92 | 57.84 | 81.08 | 10.29 | 37.86 | 12.97 | 45.95 | 41.62 | 100 |
| Klebsiella spp. | 20.83 | 54.17 | 39.17 | 58.33 | 18.18 | 19.35 | 15.83 | 38.33 | 46.67 | 100 |
| Proteus spp. | 39.13 | 34.78 | 34.78 | 43.48 | 33.33 | 25.00 | 34.78 | 56.52 | 47.83 | 100 |
| Citrobacter spp. | 42.86 | 42.86 | 42.86 | 71.43 | 33.33 | 42.86 | 57.14 | 42.86 | 85.71 | 100 |
| Pseudomonas spp. | 22.58 | 45.16 | 35.48 | 58.06 | 50.00 | 60.00 | 12.90 | 58.06 | 77.42 | 100 |
| Acinetobacter spp.* | |
Among ESBL producing genes prevalence of bla-CTX-M (82.5%) was highest, followed by bla-TEM (67.5%) and bla-SHV (57.5%) [Table/Fig-7].
Total Prevalence of ESBL producing genes
| GENE → | TEM | SHV | CTX-M |
|---|
| E. coli | 60 | 55 | 80 |
| Klebsiella sp. | 75 | 60 | 85 |
| Total Prevelance | 67.5 | 57.5 | 82.5 |
Among 20 ESBL positive strains of E. coli all the three genes i.e. bla-TEM, bla-SHV and bla-CTX-M were present in four strains of E. coli while, only two genes i.e. bla-TEM and bla-SHV were present only in one strain, bla-TEM and bla-CTX-M were present in five strains and bla-SHV and bla-CTX-M were present in three strains of E. coli. However, frequency of prevalence of single gene for ESBL was only one for bla-TEM and bla-SHV while, three for bla- CTX-M [Table/Fig-8].
Prevalence of β-Lactamase encoding genes bla-TEM, bla-SHV and bla-CTX-M in Escherichia coli
| S. No. | TEM | SHV | CTX-M |
|---|
| 1 | - | - | + |
| 2 | - | - | + |
| 3 | - | + | + |
| 4 | + | + | + |
| 5 | + | + | + |
| 6 | + | - | - |
| 7 | - | + | + |
| 8 | + | + | - |
| 9 | + | - | + |
| 10 | - | + | - |
| 11 | + | + | + |
| 12 | + | - | + |
| 13 | + | + | + |
| 14 | - | + | + |
| 15 | + | + | - |
| 16 | + | - | + |
| 17 | + | - | + |
| 18 | - | + | + |
| 19 | - | - | + |
| 20 | + | - | + |
Among 20 ESBL positive strains of Klebsiella spp. all the three genes i.e. bla-TEM, bla-SHV and bla-CTX-M were present in nine strains of Klebsiella spp. while only two genes i.e. bla-TEM and bla-SHV were present only in one strain, bla-TEM and bla-CTX-M were present in four strains and bla-SHV and bla-CTX-M were present in one strain only. However, frequency of single gene for ESBL production was only one for bla-TEM and two for bla-CTX-M. In strain no. 30 none of the gene was present [Table/Fig-9].
Prevalence of β-Lactamase encoding genes bla-TEM, bla SHV and bla-CTX-M in Klebsiella spp.
| No. | TEM | SHV | CTX-M |
|---|
| 21 | + | - | + |
| 22 | + | + | + |
| 23 | + | + | + |
| 24 | - | + | + |
| 25 | + | - | - |
| 26 | - | - | + |
| 27 | + | + | - |
| 28 | + | + | + |
| 29 | + | + | + |
| 30 | - | - | - |
| 31 | + | + | + |
| 32 | + | + | + |
| 33 | + | - | + |
| 34 | - | - | + |
| 35 | - | + | + |
| 36 | + | - | + |
| 37 | + | - | + |
| 38 | + | + | + |
| 39 | + | + | + |
| 40 | + | + | + |
Discussion
ESBLs have become a widespread serious problem. These enzymes are becoming increasingly expressed by many strains of pathogenic bacteria with a potential for dissemination. Presence of ESBL compromise the activity of wide-spectrum antibiotics creating major therapeutic difficulties with a significant impact on the outcome of patients. The continued emergence of ESBLs presents diagnostic challenges to the clinical microbiology laboratories.
In the present study ESBL production was found to be 52.49%. On the contrary in various other studies ESBL production rate varies from 17% to 70% [12–17]. In present study respiratory tract samples (63.83%) were the major source of ESBL producing strains followed by stool samples, urine, body fluid, pus and blood. However, in other studies urine was the major source of ESBL producers [13,18,19]. One of the investigator however, reported blood as major source of ESBL producers [14].
Among various GNB isolated highest ESBL production was observed in Klebsiella spp. However, in other studies E. coli was the major ESBL producer [13,20–22]. According to Umadevi et al., in Pseudomonas spp. ESBL production is less as compared to Enterobacteriaceae and is in accordance with present study [22].
Ali et al., reported ESBL production in Acinetobacter baumanii to be 72%, Proteus mirabilis to be 61%, Proteus vulgaris to be 50% which is quite high as compared to present study but ESBL production in Pseudomonas aeruginosa and Citrobacter freundii was 36.36% and is comparable with present study [16].
In present study high prevalence of ESBL producing enterobacteriaceae among hospitalized patients was observed and is in agreement with findings of other investigators [14,20].
In the present study all the ESBL isolates were sensitive to imipenam. Among β-Lactam/ β-Lactam inhibitor drugs highest sensitivity was observed with cephoparazone/sulbactum in case of E. coli (45.95%) and Proteus spp (56.52%), while with piperacillin/tazobactum in case of Klebsiella spp. (46.67%), Citrobacter spp (85.71%) and Pseudomonas spp. (77.42%) while high resistance was observed with amoxyclav. Pseudomonas spp. showed 60% sensitivity to ofloxacin and 50% to ciprofloxacin, while high resistance to quinilones was observed in enterobacteriaceae isolates. High resistance was observed with cefepime in all isolates. In the present study all the ESBL isolates were found to be Multi Drug Resistant (MDR). These findings are in agreement with other studies [13, 20–23].
Phenotypic tests for ESBL detection only confirm whether an ESBL is produced but cannot detect the ESBL subtype. Some ESBLs may fail to reach a level to be detected by disk diffusion tests but result in treatment failure in the infected patient. Nuesch & Hachler reported that although molecular methods appear sensitive, but are expensive, time consuming and require specialized equipment and expertise [24]. However, definitive identification is possible only by molecular detection methods.
There are so many types of ESBLs like TEM, SHV, CTX, OXA, AmpC, etc. but majority of the ESBLs are derivatives of TEM or SHV or CTX-M enzymes and these enzymes are most often found in E. coli and K. pneumoniae. Keeping in view this scenario, the current study was investigated upon E. coli and K. pneumoniae to look for the presence of TEM or SHV or CTX-M gene.
In present study high frequency of ESBL positive strains were observed. 195(57.18%) strains of E. coli and 108(67.08%) of were shown to produce ESBLs as investigated by disc diffusion test. Out of which 20 strains each of E. coli and Klebsiella spp. were selected randomly for molecular characterization.
A pair of forward and reverse primers was used to amplify TEM, SHV and CTX-M genes. Of 20 ESBL positive E. coli isolates, 60% harbored TEM gene, 55% harbored SHV gene and 80% harbored CTX-M gene as detected by PCR and of the 20 ESBL positive Klebsiella spp. isolates 75% harbored TEM gene, 60% harbored SHV gene and 85% harbored CTX-M gene. Jyoti et al., reported 60% TEM gene and 72% SHV gene in Klebsiella spp. and 52% TEM gene and 48% SHV gene in E. coli in her study [25]. Higher prevalence of TEM gene in Klebsiella spp. than E. coli is in accordance with present study, but however prevalence of SHV gene in Klebsiella spp. in present study was low [25].
According to another study by Bali et al., TEM type ESBLs were found in 72.72% of E. coli and 75% of Klebsiella spp., while they reported 9.09% of SHV gene and 22.72% of CTX-M gene in their study which is quite low as compared to present study [26].
The high prevalence of CTX-M gene in our study is in concordance with study of Vaida et al., who reported CTX-M-encoding genes in the majority of E. coli (96 %) and K. pneumoniae (71 %) isolates showing the ESBL phenotype [27]. Livermore et al., Bonnet stated in separate studies that the CTX-M gene is the most prevalent ESBL-encoding gene worldwide and is replacing TEM and SHV types as the predominant ESBL in many European and Asian countries [28,29].
According to King –Ting Lim et al., majority of the ESBL-positive isolates from Malaysia harbored TEM-1 (88%), which is quite high as compared to this study, but they reported prevalence of CTX-M (20%) and SHV (8%) which is very low as compared to this study [30]. Mubarak et al., highlighted the emergence and dissemination of CTX-M-15 producing E. coli and K. pneumoniae in the UAE as majority of the strains 199 (87%) in their study expressed the CTX-M gene which is in concordance with present study but the SHV gene was detected in 29 (13%) of the strains by them which is quite low as compared to this study [15].
In a study from Thailand Pattarachai Kiratisin et al., reported 99.6% of ESBL producing E. coli and 99.2% of ESBL-producing K. pneumonia isolates carried bla CTX-M and demonstrated that CTX-M-type ESBL is highly endemic in Thailand [31]. The bla TEM and bla SHV groups were detected in 77.0% and 3.8% of ESBL-producing E. coli and 71.7% and 87.4% of ESBL-producing K. pneumoniae, respectively.
In another study from Turkey the most frequent β-lactamase type was CTX-M (92%), followed by TEM (39%), SHV (5%) and Vietnamese extended-spectrum beta-lactamase (VEB, 1.6%) [32].
According to Goyal et al., majority of strains (57.3%) harbored 2 or more ESBL genes, while Bali et al., [26,33] observed that 19.2% ESBL isolates carried more than one type of β lactamase genes. In the present study also it has been observed that 31 out of 40 (77.5%) isolates carried more than one type of β lactamase genes which is quite high as compared to previous studies, with 13 (32.5%) isolates harboring all the three β lactamase genes, 9 (22.5%) isolates harboring TEM and CTX, 6 (15%) isolates harboring SHV and CTX and 3 (7.5%) isolates harboring TEM and SHV.
ESBL strains are usually multi-drug resistant. Because these strains become resistant to available antibiotics and they can pass the gene to other clinical strains, the quick detection of these strains in microbiology laboratories is very important. Antimicrobial therapy has played an important role in the treatment of human bacterial infections, but the drug resistance that has emerged in the treatment of bacterial infections due to ESBL enzymes degrades all beta lactam antibiotics and thus bacteria become multidrug resistant [34].
Conclusion
The present study highlights the prevalence of ESBL-producing bacteria in the rural area of Jaipur, Rajasthan, India. All the strains isolated were multi drug resistant and retained their sensitivity against imipenam. In view of this emerging drug resistance the practice of routine ESBL testing along with conventional antibiogram would be useful for all cases which will help in the proper treatment of the patient and also prevent further development of bacterial drug resistance. The study reveals high prevalence of CTX-M gene in our hospital. Molecular typing would determine which types of ESBL are present in each isolate. Molecular detection and identification of beta lactamases would be essential for a reliable epidemiological investigation of antimicrobial resistance. These enzymes can be chromosomal or plasmid mediated, which may help in the dissemination of antimicrobial drug resistance in health care settings. Therefore, ESBL producing organisms should be identified quickly so that appropriate antibiotic usage and infection control measures can be implemented.