Background & Objectives: Myocardial infarction and stroke are leading causes of death worldwide. Primarily, arteriosclerosis is responsible for these events. There is a strong family history suggesting a genetic cause. Apolipoprotein E (apo E) plays an important role in lipid metabolism. Apo E is polymorphic with three isoforms, ApoE2, ApoE3 and ApoE4; which translate into three alleles of the gene. Its polymorphism may be a risk determinant of atherosclerosis.
Methods: Lipoprotein concentrations were studied, in 100 myocardial infarction and 50 cerebrovascular stroke subjects and compared with age and sex matched controls. Genotypes for apo E isoforms (E2, E3, and E4) for all above subjects and age and sex matched controls were determined by Multiplex Amplification Refractory Mutation System PCR.
Results: There were statistically significant higher values of serum total cholesterol and LDL cholesterol in study group, as compared to control group. Study of Apo E isoforms revealed higher proportion of E4 allele in the study group as compared to control group. The occurrence of each allele frequency in study and control group was E4E4: 28.66% and 16.0%, E3E3: 39.33% and 56.66%, E4E3: 14.66% and 9.33%, E3E2: 8.66% and 10.66%, E4E2: 4.66% and 2.66% & for E2E2: 4.0% and 4.66% respectively.
Interpretation & Conclusion: There were significantly higher levels of serum total cholesterol, LDL cholesterol and triglyceride with E4 allele; when compared with in the study group and between study group and control group. Apo E polymorphism influences serum lipid levels and is an independent risk determinant of arteriosclerosis.
Apolipoprotein E, Arteriosclerosis, Cerebrovascular Stroke, Coronary artery disease, Hyperlipidemia, Myocardial infarction
Introduction
Myocardial infarction and stroke are leading causes of death worldwide[1]. Lifestyle and environmental factors play an important role in their development [2]. Myocardial Infarction or stroke is a medical emergency and can cause damage to myocardial muscle or permanent neurological damage and death. Risk factors for stroke include old age, high blood pressure, previous stroke or transient ischaemic attack, diabetes, high cholesterol, tobacco smoking & chewing and arterial fibrillation. High blood pressure is the most important modifiable risk factor of arteriosclerosis [3].
Apolipoprotein E (apo E) is a serum glycoprotein consisted of 299 amino acids found in circulating chylomicrons, chylomicron remnants, Very Low Density Lipoproteins (VLDL), Intermediate Density Lipoproteins (IDL) and high-density lipoproteins (HDL)4. Apo E gene consists of four exons and three introns spanning 3,597 nucleotides and produces a 299 amino acid polypeptide [4,5]. It is synthesized primarily in the liver, but other organs and tissues also synthesize apo E, including brain, spleen, kidneys, gonads, adrenals, and macrophages [6].
The structural gene locus for plasma apo E is polymorphic having three common alleles, designated as E2, E3 and E4. Consequently, three homozygous (E2/E2, E3/E3, E4/E4) and three heterozygous (E3/E2, E4/E2 and E4/E3) phenotypes are found in the general populationn[7]. The product of the three alleles differ in properties such as its affinity for binding to apo E and low-density lipoprotein receptors (LDL-R), and its affinity for lipoprotein particles [8].
For decades, Apo E has been regarded as one of the important factor of lipoprotein genetics [9,10]. Several studies have demonstrated the impact of Apo E polymorphisms in cerebrovascular and cardiovascular diseases in a reproducible fashion. The Apo-E isoforms have consistently been shown to be associated with variation in plasma LDL cholesterol level, with the E4 allele exerting a greater positive influence than E3. Apo-E has consistently shown significant gene - environment interactions, modulating its association with plasma lipid parameters as well as cardiovascular and cerebrovascular risk [11,12].
The aim of this study was to demonstrate the influence of the Apo-E gene polymorphism on plasma lipid levels which is an important determinant of myocardial infarction and cerebrovascular stroke in the study population group of Western Maharashtra,India. It was also aimed to elucidate role of Apo-E gene polymorphism in the study population as a risk factor alongwith traditional risk factors.
Material and Methods
Subject
The present study was carried out in the Department of Biochemistry, Government Medical College, Miraj and Department of Biochemistry, BJ Government Medical College, Pune. Approval was obtained from the institutional ethical committee of Government Medical College, Miraj, India.
A total of 300 subjects which consist of 150 Arteriosclerosis patients and 150 age & sex matched controls were recruited for the study from Medicine Department. Out of the 150 arteriosclerosis patients, 100 were myocardial infarction patients and 50 were cerebrovascular stroke patients. Clinical diagnosis of myocardial infarction and cerebrovascular stroke was confirmed by relevant diagnostic modalities such as electrocardiogram, or CT Scan studies of brain, along with routine biochemical investigations. The informed consent was obtained from all subjects for participation in the study.
Venous Blood samples were collected from patients after an overnight fast in tube containing EDTA, placed on crush ice, spun immediately, and plasma was separated. Plasma samples were analyzed for total Cholesterol, HDL Cholesterol & triglyceride. Plasma total Cholesterol, triglyceride, HDL Cholesterol concentrations were quantified using an automated clinical chemistry analyzer.
Serum Total Cholesterol levels was estimated by enzymatic method [13]. For HDL cholesterol estimation, Chylomicrons, VLDL cholesterol and LDL cholesterol fractions in plasma were separated from HDL cholesterol by precipitating with phosphotungstic acid and magnesium chloride. After centrifugation the cholesterol in the HDL fraction, which remains in the supernatant, was assayed with enzymatic cholesterol method [13]. LDL cholesterol level was calculated by using Friedewald formula [14].
Serum triglyceride was determined by Glycerol phosphate oxidase-peroxidase method [15]. The results were expressed as mean ± SD and was analyzed statistically.
Genetic Analysis
Either fresh or frozen venous blood samples in EDTA were subsequently used for DNA extractions by salting out method [16]. Genotypes for Apo-E isoforms (E2, E3, and E4) for all the above subjects and controls were determined by Multiplex Amplification Refractory Mutation System PCR [17].
The mutant & wild specific ARMS primers, control primers, and common primer used to diagnose Apo E polymorphisms in this study which are listed in [Table/Fig-1]. For each reaction there were total five primers used, two of these five primers serve as internal control. A common primer was designed that paired with Arg/Cys 158 or Arg/Cys 112 and produced an amplicon of 588 and 451 base pairs (bp), respectively. A 360-bp fragment of the a1-antitrypsin gene was coamplified to function as an internal control [18,19].
The mutation-specific ARMS primers, control primers, and common primers for Apo-E polymorphism
| Sr. No. | Mutation/wild specific/common/control primer | Primer sequence (5’–3’) | Length of PCR product (bp) |
|---|
| 1 | Cys158 | ATGCCGATGACCTGCAGAATT | 588 |
| 2 | Arg158 | ATGCCGATGACCTGCAGAATC | 588 |
| 3 | Cys112 | CGCGGACATGGAGGACGTTC | 451 |
| 4 | Arg112 | CGCGGACATGGAGGACGTTT | 451 |
| 5 | COMMON | GTTCAGTGATTGTCGCTGGGCA | - |
| 6 | CONTROL FOR | CCCACCTTCCCCTCTCTCCAGGCAAATGGG | 360 |
| 7 | CONTROL RES | GGGCCTCAGTCCCAACATGGCTAAGAGGTG |
The PCR was carried out in a total volume of 25 μl of reaction mixture, containing 50 ng of genomic DNA; 12.5 μl 2X master mix containing 200 mmol/L of deoxynucleotides triphosphates; 10 mmol/L Tris-HCL, pH 8.8; 1.5 mmol/L MgCl2; 50 mmol/L KCl, and 1 mL/L Triton X-100; 2.5 U of Taq DNA Polymerase; and then add 80 g/L dimethyl sulfoxide (DMSO; Sigma); 8 nmol/L of control primers, and 0.8 mmol/L ARMS common primer. Reaction mixture A contained, in addition to the above,0.8 mmol/L Arg158 and 0.4 mmol/L Arg112 primers. Similarly, reaction mixture B contained, in addition to the above, 0.8 mmol/L Cys158 and 0.4 mmol/L Cys112 primers. Amplification was performed with a Quanta Biotech DNA thermocycler. Each PCR run had blank controls with no DNA to exclude contamination. Cycling was carried out on the thermal cycler with initial denaturation at 95°C for 4 minutes, 35 cycles with denaturation at 94 °C for 45 second, annealing at 66 °C for 45 s, and extension at 72 °C for 45 second, followed by a final cycle of extension at 72 °C for 5 minutes.
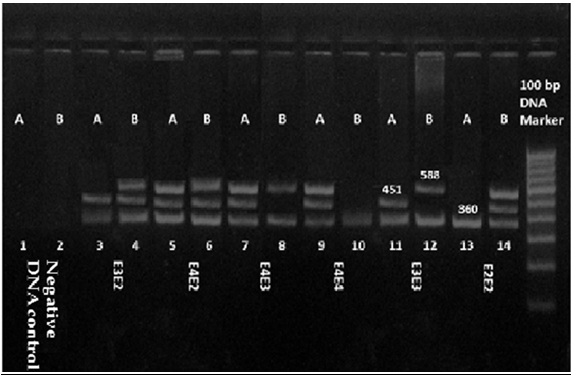
10 μl amplified product was mixed with 2 μl of gel loading dye and separated in a 2% agarose gel that contained 0.1 mg/L ethidium bromide. The samples were electrophoresed for about 1 hour at 100 volts, using 0.5 X Tris-Acetate-EDTA running buffer. The amplicons were sized by using a 100-bp DNA marker. Amplification products of the six common Apo E genotypes with modified ARMS on agarose gel electrophoresis are shown in [Table/Fig-2].
Amplification products of the six common Apo E alleles with modified ARMS on agarose gel electrophoresis. Each sample was analyzed by loading two different PCR reaction mixtures. Mixture A contains the Arg112 (451 bp) and Arg158 (588 bp) specific ARMS primers. Similarly, mixture B contains the specific ARMS primers Cys112 (451 bp) and Cys158 (588 bp). Both mixtures contain the same common ARMS primer as well as a set of internal control primers that amplify a 360-bp segment of the α1-antitrypsin gene.
Lanes 1 and 2, negative DNA control;
Lane 3 and 4, E3E2(Arg112 positive, Arg158 negative; Cys112 positive, Cys158 positive);
Lane 5 and 6, E4E2 (Arg112 positive, Arg158 positive; Cys112 positive, Cys158 positive);
Lane 7 and 8, E4E3(Arg112 positive, Arg158 positive; Cys112 negative, Cys158 positive);
Lane 9 and 10, E4E4(Arg112 positive, Arg158 positive; Cys112 negative, Cys158 negative);
Lane 11 and 12, E3E3 (Arg112 positive, Arg158 negative; Cys112 negative, Cys158 positive);
Lane 13 and 14, E2E2 (Arg112 negative, Arg158 negative; Cys112 positive, Cys158 positive);
Lane 15, 100 bp DNA marker
Discussion
All enrolled subjects were recruited from South Western Maharashtra region,India. The data of biochemical parameters in Arteriosclerosis Patients & controls were summarized in [Table/Fig-3].It was observed that arteriosclerosis patients had significantly higher plasma total cholesterol concentrations as compare to controls (p<0.0001). Arteriosclerosis individuals had significantly greater mean value for LDL Cholesterol and VLDL cholesterol (p<0.0001) than controls. The mean level of HDL Cholesterol level was significantly lower (p<0.0001) in patients of arteriosclerosis subjects than control. The arteriosclerotic group had significantly higher plasma triglyceride (p<0.0001) than the control group.
Levels of Biochemical Parameters in Arteriosclerosis Patients & controls
| Biochemical Parameters | Patients (n=150) | Controls (n=150) | p– value |
|---|
| Total Cholesterol (mg/dl) | 221.09 ± 53.58 | 173.77 ± 22.62 | <0.0001 |
| HDL Cholesterol (mg/dl) | 37.69 ± 11.31 | 48.59 ± 10.36 | <0.0001 |
| LDL Cholesterol (mg/dl) | 159.08 ± 56.07 | 106.70 ± 25.30 | <0.0001 |
| VLDL Cholesterol (mg/dl) | 24.32 ± 10.88 | 18.47 ± 5.59 | <0.0001 |
| Triglyceride (mg/dl) | 121.61 ± 54.41 | 92.35 ± 27.99 | <0.0001 |
The present investigation was aimed to examine whether significant association does exist between Apo-E gene polymorphism and arteriosclerosis in study population. [Table/Fig-4]shows Frequency Distribution of Apo-E alleles in arteriosclerosis patient and controls. The occurrence of each allele frequency in study and control group was E4E4: 28.66% and 16.0%, E3E3: 39.33% and 56.66%, E4E3: 14.66% and 9.33%, E3E2: 8.66% and 10.66%, E4E2: 4.66% and 2.66% & for E2E2: 4.0% and 4.66% respectively. We observed that the E3 allele was the most frequent, followed by the E4 allele and E2 allele. This finding is universal for human populations [20]. It was also conferred by Singh et al., [21,22], that the E3 allele and E3/ E3 alleles were most common in normal and angiographically diagnosed CHD patients. Study on patients with clinically proven coronary artery disease, show increase in plasma total cholesterol and LDL cholesterol according to the Apo E phenotype in the order APO E3/2 < E3/3<E3/4 and E4/4[23].
Frequency Distribution of Apo E alleles in Arteriosclerosis patient & controls
| Genotypes | Patients (n=150) | Controls (n=150) |
|---|
| E 2 E 2 | (06) 04.00% | (07) 04.66% |
| E 3 E 3 | (59) 39.33% | (85) 56.66% |
| E 4 E 4 | (43) 28.66% | (24) 16.00% |
| E 3 E 2 | (13) 08.66% | (16) 10.66% |
| E 4 E 2 | (07) 04.66% | (04) 02.66% |
| E 4 E 3 | (22) 14.66% | (14) 09.33% |
Association of Apo E phenotypes with lipid profile in Arteriosclerosis patients & controls were summarised in [Table/Fig-5]. It was observed that in arteriosclerosis patients having homozygous E2 allele have no statistical difference in their means of total cholesterol, HDL cholesterol, LDL cholesterol, VLDL cholesterol and triglycerides than normal healthy controls with homozygous E2 allele. Homozygous E3 allele individuals had significantly higher mean value for Total Cholesterol (p<0.0001), LDL Cholesterol (p<0.0001), VLDL cholesterol (p = 0.0027) and triglyceride (p=0.0027), and significantly lower HDL Cholesterol (p<0.0001) was found in patients than control of the same genotype. The mean levels of Total Cholesterol (p<0.0001), LDL Cholesterol (p<0.0001), VLDL cholesterol (p= 0.0001) and triglycerides (p= 0.0001) were significantly increased and HDL Cholesterol (p= 0.0196) level was decreased among patients of arteriosclerosis subjects group than control subject in homozygous E4 allele. The arteriosclerotic group with E3E2 allele had statistically higher plasma Total Cholesterol (p= 0.0003), LDL Cholesterol (p= 0.0013) and no variation was found in HDL Cholesterol (p= 0.3481), VLDL cholesterol (p= 0.1469) and triglyceride (p = 0.1469) levels than the normal control group. E4E2 allele individuals had significant decreased level of HDL Cholesterol (p = 0.0423), and there was no significant difference found for Total Cholesterol (p = 0.1869), LDL Cholesterol (p = 0.0995), VLDL cholesterol (p = 0.8306) and triglycerides (p = 0.8306) in patients than controls. Arteriosclerotic Heterozygous E4E3 allele individuals have significantly higher mean value for Total Cholesterol (p = 0.0029) and LDL Cholesterol (p = 0.0101), no significant difference was found in HDL Cholesterol (p = 0.1644), VLDL cholesterol (p = 0.1013) and triglyceride (P = 0.1013) than control E4E3 allele individuals. Our result shows homozygous E4 alleles with higher plasma lipid levels, in the study as well as control group [Table/Fig-4]. Apo E4 allele has been linked to higher total cholesterol, LDL cholesterol, VLDL cholesterol and triglycerides and lower HDL cholesterol plasma levels in several studies[24].
Association of Apo E phenotypes with lipid profile in Arteriosclerosis patients & controls
| Apo E alleles | Subjects | Total Cholesterol (mg/dl) | HDL Cholesterol (mg/dl) | LDL Cholesterol (mg/dl) | VLDL Cholesterol (mg/dl) | Triglycerides (mg/dl) |
|---|
| E2E2 | Patients | 132.5 ± 19.02 | 36 ± 7.48 | 73.8 ± 20.08 | 22.7 ± 6.77 | 113.5 ± 33.85 |
| Controls | 153.84 ± 17.33 | 46.61 ± 11.77 | 88.05 ± 13.19 | 19.17 ± 5.87 | 95.86 ± 29.36 |
| E3E3 | Patients | 214.29 ± 51.41* | 35.59 ± 9.13* | 155.80 ± 56.12* | 22.90 ± 12.45* | 114.48 ± 62.23* |
| Controls | 167.08 ± 18.96 | 50.02 ± 10.42 | 98.88 ± 21.54 | 18.18 ± 5.72 | 90.92 ± 28.62 |
| E4E4 | Patients | 252.63± 55.40* | 38.61 ± 13.49* | 186.69 ± 57.22* | 27.33 ± 9.23* | 136.66 ± 46.15* |
| Controls | 198.50 ± 21.24 | 46.35 ± 11.10 | 133.40 ± 22.86 | 18.74 ± 4.56 | 93.71 ± 22.80 |
| E3E2 | Patients | 199.46 ± 18.41* | 44.54 ± 14.56 | 132.12 ± 19.27* | 22.80 ± 11.73 | 114.00 ± 58.67 |
| Controls | 170.50 ±18.66 | 48.71 ± 8.73 | 103.82 ± 22.39 | 17.97 ± 4.97 | 89.87 ± 24.84 |
| E4E2 | Patients | 230.71 ± 50.91 | 32.29 ± 9.64* | 180.31 ±56.61 | 18.11 ± 4.67 | 90.57 ± 23.34 |
| Controls | 193.25 ± 8.30 | 52.00 ± 18.58 | 123.85 ± 28.49 | 17.40 ± 6.06 | 87.00 ± 30.31 |
| E4E3 | Patients | 211.62 ± 33.03* | 39.67 ± 9.77 | 146.36 ± 37.45* | 25.58 ± 10.26 | 127.92 ± 51.31 |
| Controls | 180.21 ± 19.46 | 43.70 ± 5.06 | 116.25 ± 21.63 | 20.27 ± 7.27 | 101.34 ± 36.34 |
* Stastically significant
Comparison of the lipid values with patients and within the control group having E4 homozygous allele (Group M) and alleles other than E4 homozygous allele (Group N) was done. We found statistically significant higher values of total cholesterol (p < 0.0001), LDL cholesterol (p = 0.0001) in group M patients than Group N patients. The difference in VLDL cholesterol (p= 0.0313) & triglyceride level (p = 0.0313) was found to be significantly higher in group M than in Group N in patients. There was no significant change found in HDL cholesterol (p = 0.5298) in Group M patients and Group N.
The similar findings were seen regarding serum cholesterol (P < 0.0001), LDL cholesterol (P < 0.0001) and HDL cholesterol (P = 0.2487) in group M and group N within the control group. And in contrast to patient group, there was no significant change for VLDL cholesterol (P = 0.7964) & triglycerides (P = 0.7964) in group M and group N within control group. The study suggested that the E4 homozygous allele affects plasma cholesterol and LDL cholesterol levels and hence, it is an independent risk factor for the development of arteriosclerosis.
The prevalence of the E4-containing phenotypes was significantly higher in cardiovascular and cerebrovascular ailments such as myocardial infarction, hypertension, coronary heart disease and stroke etc [25]. Mendes-Lana A demonstrated that the presence of Apo E2 can confer a protective effect, the presence of Apo E4 implies an enhanced risk of dyslipidemia [26], and our results are in accordance with above studies for E 4 allele. The study confirms the significant association between ApoE4 allele and hyperlipidemia.
Since, the population in the state of Maharashtra,India is divided into different caste and tribal groups and there is very less in breeding, it is necessary to study the representative sample from each caste group so as to assess the extent and impact of Apo E polymorphism in South Western Maharashtra population. This will enlighten the understanding of the association between Apo-E polymorphism and Hyperlipidemia among the study population.
Conclusion
Study supports that variability in Apo-E gene locus is associated with influencing the plasma lipid levels which are important risk factors for arterial disease, and it is necessary to carry out a caste group-wise randomised study to assess the impact of Apo-E polymorphism in this population.
* Stastically significant
[1]. Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ, Global and regional burden of disease and risk factors, 2001: systematic analysis of population health dataLancet 2006 367:1747-57. [Google Scholar]
[2]. Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control studyLancet 2004 364:937-52. [Google Scholar]
[3]. Donnan GA, Fisher M, Macleod M, Davis SM, StrokeLancet 2008 371(9624):1612-23. [Google Scholar]
[4]. Scott J, Knott TJ, Shaw DJ, Brook JD, Localization of genes encoding Apolipoprotein CI, CII, and E to the p13->cen region of human chromosome 19Hum Genet 1985 71:144-46. [Google Scholar]
[5]. Rall SC, Weisgraber KH, Mahley RW, Human Apolipoprotein E: the complete amino acid sequenceJ Biol Chem 1982 257:4171-78. [Google Scholar]
[6]. Mahley RW, Apolipoprotein E: cholesterol transport protein with expanding role in cell biologyScience 1988 240:622-30. [Google Scholar]
[7]. Ulrike B, Wilfried W, Gudrun I, Joachim H, Keith KS, The LDL- receptor- related protein LRP is an Apolipoprotein E binding proteinNature 1989 341:162-64. [Google Scholar]
[8]. Davignon J, Gregg RE, Sing CF, Apolipoprotein E polymorphism and atherosclerosisArteriosclerosis 1988 8:1-21. [Google Scholar]
[9]. Visvikis-Siest S, Marteau JB, Genetic variants pre disposing to cardiovascular diseaseCurr Opin Lipidol 2006 17:139-51. [Google Scholar]
[10]. Blackman JA, Worley G, Strittmatter WJ, Apolipoprotein E and brain injury: implications for childrenDevelop Med Child Neurol 2005 47:64-70. [Google Scholar]
[11]. Buttini M, Orth M, Bellosta S, Akeefe H, Pitas RE, Wyss-Coray T, Expression of human Apolipoprotein E3 or E4 in the brains of APO e-/- mice: isoform specific effects on neurodengerationJ Neurosci 1999 19:4867-80. [Google Scholar]
[12]. Wilson PWF, Myers RH, Larson MG, Ordovas JM, Wolf PA, Schaefer EJ, Apolipoprotein E alleles, Dyslipidemia and coronary heart disease. The Framingham offspring studyJAMA 1994 272:1666-71. [Google Scholar]
[13]. Allain CC, Poon LS, Chan CS, Richmond W, Fu PC, Enzymatic determination of total serum cholesterolClin Chem 1974 20(4):470-75. [Google Scholar]
[14]. Friedewald WT, Levy RI, Fredrickson DS, Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifugeClin Chem 1972 18:499-502. [Google Scholar]
[15]. Bucolo G, David H, Quantitative determination of serum triglycerides by the use of enzymesClin Chem 1973 19(5):476-82. [Google Scholar]
[16]. Miller SA, Dykes DD, Polesky HF, A simple salting out procedure for extracting DNA from human nucleated cellsNucleic Acids Res 1988 16(3):1215 [Google Scholar]
[17]. Donohoe GG, Salomäki A, Lehtimäki T, Pulkki K, Kairisto V, Rapid Identification of Apolipoprotein E Genotypes by Multiplex Amplification Refractory Mutation System PCR and Capillary Gel ElectrophoresisClin Chem 1999 45(1):143-46. [Google Scholar]
[18]. Wenham PR, Newton CR, Price WH, Analysis of Apolipoprotein E genotypes by the Amplification Refractory Mutation SystemClin Chem 1991 37:241-44. [Google Scholar]
[19]. Newton CR, Kalsheker N, Graham A, Powell S, Gammack A, Riley J, Diagnosis of alpha 1-antitrypsin deficiency by enzymatic amplification of human genomic DNA and direct sequencing of polymerase chain reaction productsNucleic Acids Res 1988 16:8233-43. [Google Scholar]
[20]. Burman D, Mente A, Hegele RA, Islam S, Yusuf S, Anand SS, Relationship of the ApoE polymorphism to plasma lipid traits among South Asians, Chinese, and Europeans living in CanadaAtherosclerosis 2009 203:192-200. [Google Scholar]
[21]. Singh PP, Singh M, Bhatnagar DP, Kaur TP, Gaur SK, Apolipoprotein E polymorphism and its relation to plasma lipids in coronary heart diseaseIndian J Med Sci 2008 62:105-12. [Google Scholar]
[22]. Singh PP, Singh M, Mastana SS, Apo E distribution in world populations with new data from India and the U.K.Ann Hum Biol 2006 33:279-308. [Google Scholar]
[23]. Shajith A, Misra A, Meena K, Luthra K, Apolipoprotein E polymorphism in cerebrovascular & coronary heart diseasesIndian J Med Res 2010 132:363-78. [Google Scholar]
[24]. Haddy N, De Bacquer D, Chemaly MM, Maurice M, Ehnholm C, Evans A, The importance of plasma Apolipoprotein E concentration in addition to its common polymorphism on inter-individual variation in lipid levels: results from Apo EuropeEur J Hum Genet 2002 10: 841-50. [Google Scholar]
[25]. Lehtinen S, Lehtimäki T, Sisto T, Salenius JP, Nikkilä M, Apolipoprotein E, serum lipids, myocardial infarction and severity of angiographically verified coronary artery disease in men and womenAtherosclerosis 1995 114:83-91. [Google Scholar]
[26]. Mendes-Lana A, Pena GG, Freitas SN, Lima AA, Nicolato RL, Nascimento-Neto RM, Apolipoprotein E polymorphism in Brazilian dyslipidemic individuals: Ouro Preto StudyBraz J Med Biol Res 2007 40:49-56. [Google Scholar]