Approval of the institutional ethics committee was obtained for the conduct of this study.
Methods
Susceptibility of these organisms to aztreonam (30μg) , cefotaxime (30 μg), ceftazidime (30μg), cefepime (30 μg ), piperacillin-tazobactam (100/10 μg ), ciprofloxacin (5 μg), amikacin(30 μg), ertapenem(10 μg), imipenem (10 μg) and meropenem(10 μg) (Hi-Media Laboratories, India) was tested by the disc diffusion method and results were interpreted as per CLSI 2011 guidelines [10]. Susceptibility of these organisms to tigecycline was determined using 15μg tigecycline disc (BBL TM BD,USA). The criteria of the United States, Food and Drug Administration, was used for interpretation [11].
Minimal Inhibitory Concentrations (MICs) to imipenem and meropenem were determined by broth microdilution method and results were interpreted according to CLSI document M100-S21. MIC to colistin was determined by E test (Biomerieux SA, France) [10].
Production of Carbapenemase was screened by the Modified Hodge test (MHT) [10].
Polymerase chain reaction (PCR): All study isolates were subjected to PCR by using primers which targeted bla OXA-181 and bla OXA-48.
The primers which were used were: OXA- 181 forward seq: ATGCGTGTATTAGCCTTATCG; OXA-181 reverse seq: AACTACAAGCGCATCGAGCA; [5] and OXA-48 like forward seq: GCTTGATCGCCCTCGATT; OXA-48 like reverse seq: GATTTGCTCCGTGGCCGAAA [12].
DNA sequencing: PCR products of the isolates that carried the bla OXA-181 gene were purified by using DNA purification kit (QIA quick Gel Extraction Kit , Qiagen ,Valencia, CA, USA) and they were subjected to automated DNA sequencing (ABI 3100, Genetic Analyser, Applied Biosystems, Foster city, CA, USA). The aligned sequences were analysed using the Bioedit sequence program and similarity searches for the nucleotide sequences were performed using the BLAST program (http://www.ncbi.nlm.nih.gov).
Results
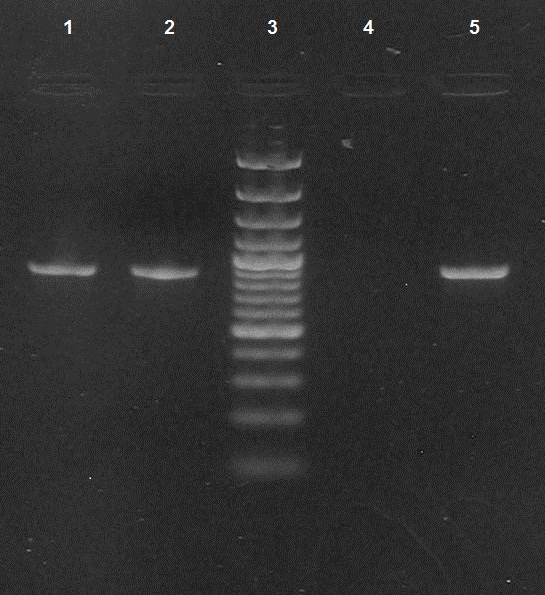
Two isolates were found to be positive for OXA-48 and OXA-181 by PCR. The PCR screening results were validated by sequencing and the sequences of the bla oxa-181 genes showed 100% identity with previously reported genes [Table/Fig-1]. Shows the results of PCR for detection of bla OXA-181. These were Klebsiella pneumoniae and Citrobacter freundii, which were isolated from patients who were admitted to the Intensive Care Units (ICU). While Klebsiella pneumoniae was isolated from a patient with septicaemia, Citrobacter freundii was isolated from the urine of a catheterised patient with multiple injuries and had undergone a neurosurgical procedure. MICs to imipenem and meropenem for Klebsiella pneumoniae were 128mg/l and 64 mg/l and for Citrobacter freundii, were was 32mg/l and 16mg/l respectively. The Modified Hodge test revealed that both were strong carbapenemase producers. The OXA-181 producers were susceptible only to tigecycline and colistin (MIC- 0.38mg/L) and were resistant to cefotaxime, ceftazidime, aztreonam, amikacin, ciprofloxacin, piperacillin-tazobactam and ertapenem. [Table/Fig-2] shows the clinical characteristics of OXA-48/OXA-181 producing Klebsiella pneumoniae and Citrobacter freundii.
PCR for detection of bla OXA-181 Lanes 1and 2: bla OXA-181 positive (amplicon size- 888bp) ; Lane 3: Molecular mass marker (100bp DNA ladder); Lane 4: negative control ; Lane 5: positive control
Clinical Characteristics of the patients infected with OXA -48 producing isolates
| Characteristics | Patient -1 | Patient -2 |
|---|
| Isolate Number | MS 2218 | MS2287 |
| Isolate | Klebsiella pneumoniae | Citrobacter freundii |
| Age /sex | 73 years, female | 56 years ,male |
| Hospital location | ICU | ICU |
| Admitting unit | Cardiology | Neurosurgery |
| Underlying disease | Coronary heart disease | Road traffic accident |
| Co-morbid conditions | Diabetes mellitus | none |
| Days in hospital | 35 days | 45days |
| Surgical procedures ,if any | Coronary artery bypass grafting | Craniotomy |
| Anti–microbials used prior to detection of OXA-181 | Cefoperazone- sulbactam, vancomycin | Ceftriaxone, cefoperazone -sulbactam, amikacin, imipenem, metronidazole |
| Indwelling devices | Internal jugular venous catheter, peripheral venous line, urinary catheter | Internal jugular venous catheter, peripheral venous line, urinary catheter |
| Mechanical ventilation | Intubated | Intubated |
| Outcome | Recovered | Expired |
| Source specimen | Blood | Urine |
| MIC –IMP mg/L | 128 | 32 |
| MIC –MEM mg/L | 64 | 16 |
| MHT | Positive | Positive |
| ESBL screen test | Positive | Positive |
| Carbapenemase | OXA-48/OXA-181 | OXA-48/OXA-181 |
| Anti–microbial susceptibility profile Susceptible to | Colistin and tigecycline | Colistin and tigecycline |
| Anti–microbial susceptibility profile Resistant to | amikacin, ciprofloxacin,aztreonam, piperacillin-tazobactam and ceftazidime | amikacin, ciprofloxacin, aztreonam, piperacillin-tazobactam and ceftazidime |
Discussion
Generally, OXA-48 carbapenemase and its variants hydrolyse penicillins at a high level, but hydrolyse carbapenems at a low level only. They show very weak activity against extended spectrum cephalosporins. Hence, the bla OXA-48 producers exhibit reduced susceptibility to carbapenems .Their MIC to carbapenems may remain in the susceptible range, thus making their detection problematic [4]. In such infections, treatment with carbapenems results in adverse outcomes. Their detection is therefore crucial for institution of appropriate therapy and to initiate preventive measures [13].
In this study, bla OXA-48 and bla OXA-181 were detected in two of the 111 isolates. These were Klebsiella pneumoniae and Citrobacter freundii, isolated from patients admitted to the Intensive Care Units (ICU). The Modified Hodge test was positive in both. Since it has been reported that bla OXA-181 coexists with other carbapenemase genes [2, 5], we have screened these isolates for the concurrent presence of blaNDM-1, blaVIM, blaIMP and blaKPC. However, none of these genes were detected in them. In the SENTRY study which was conducted between 2006-07, out of a total of 1443 isolates, 39 were carbapenem resistant. Among these 39, 10 were found to harbour OXA-181 alone and one Klebsiella pneumoniae isolate carried both bla OXA-181 and bla VIM-5 [5]. Data from the SMART study of 2009, which was done on 235 Indian isolates that were non susceptible to ertapenem, indicated that bla OXA-48 existed alone (n= 3) or in combination with bla NDM-1 in Enterobacter cloacae (n=2) [2].
The indication for the presence of blaOXA-48 is often very ambiguous, because some of the isolates may be carbapenem resistant but may remain susceptible to cephalosporin subclass III, which may be regarded as the potential treatment of choice [14]. However, in the Indian scenario, this phenomenon may be encountered very rarely, because of the high prevalence of ESBLs among Enterobacteriaceae. Due to this the resistance pattern becomes broader, leaving behind only limited therapeutic options such as polymyxins and tigecycline. [14]. The OXA -181 producers in this study were ESBL screen test positive.
Unlike the observation that OXA-48/ OXA -181 had only a low level resistance to carbapenems, our isolates had MICs which were in the range of 16-128 mg/L, way above the resistance breakpoints. This may be attributed to the coexistence of ESBLs in combination with porin loss [9].
The OXA-181 producers remained susceptible only to colistin and tigecycline in-vitro. Both patients were treated with colistin. While the patient with Klebsiella pneumoniae septicaemia recovered, the other patient expired. Many patients with infections caused by OXA producers have significant comorbidity and a prolonged hospital stay, as has been observed in this study [13].
Therefore it can be reasonably assumed that production of OXA-48 /OXA-181 is not a major mechanism of carbapenem resistance. Out of 111 Enterobacteriaceae which were screened, only 2 (1.8%) were found to be OXA-181 producers. This was in sharp contrast to the findings of study of Castanheira et al., where of 39 Enterobacteriaceae, 10 were OXA-181 producers [5] and in the SMART study, among 66 isolates with a carbapenemase encoding gene, 5 were found to be blaOXA-48 positive [2]. Additionally, bla OXA-48 / blaOXA-181 in our study did not coexist with other carbapenemase encoding genes, as was reported in the earlier studies [2,5]. Notably, the carbapenem MICs were high. Since there are no specific phenotypic tests for the detection of this enzyme, PCR is the gold standard for their routine identification in clinical microbiology laboratory.