Material and Methods
Bacterial Isolates
From August 2011 to July 2012, this study was conducted on a total of two hundred consecutive non duplicate isolates of P. aeruginosa which were isolated from different clinical specimens such as urine, pus, blood and body fluids from the patients who were admitted to a 1000 bedded tertiary care hospital in Pune, India. All the specimens were collected by using strict aseptic precautions and they were immediately processed without any delay. The bacterial identification was performed by routine conventional microbial culture and biochemical tests using standard recommended techniques [5]. Further identification was done by using the VITEK-2 System (bioMérieux, Marcy l’Etoile, France).
Antimicrobial Susceptibility Testing
The antibiotic sensitivity test was performed by the standard Kirby Bauer disc diffusion technique, using commercially available discs (Hi Media, Mumbai, India) on Mueller Hinton agar plates. The antibiotics which were tested were as follows (potency in μg/disc): Piperacillin (100μg), Ticarcillin (75μg), Piperacillin-Tazobactam (100/10μg), Ticarcillin-Clavulanic acid (75/10μg), Ceftazidime (30μg), Cefepime (30μg), Aztreonam (30μg), Imipenem (10μg), Meropenem (10μg), Colistin (10μg), Polymyxin B(300 units), Gentamicin (10μg), Tobramycin (10μg), Amikacin (30μg), Netilmicin (30μg), Ciprofloxacin (5μg), Levofloxacin (5μg), Lomefloxacin (10μg) and Ofloxacin (5μg). The diameter of the zone of inhibition for each antibiotic was measured and it was interpreted as resistant, intermediate susceptible or susceptible, as per the guidelines of the Clinical Laboratory Standards Institute (CLSI) [6]. The E. coli ATCC 25922 and the P. aeruginosa ATCC 27853 strains were used for the quality control.
The minimum inhibitory concentrations (MICs) of the antibiotics were determined using VITEK-2 (bioMérieux, Marcy l’Etoile, France) and the results are given in [Table/Fig- 1].
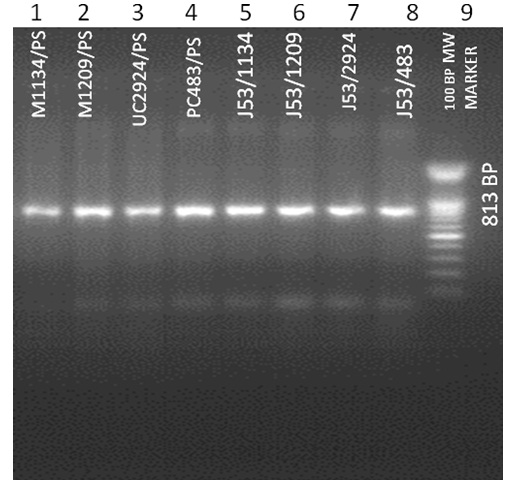
813 bp blaNDM-1 gene in P. aeruginosa isolates and their transconjugants E. coli J 53.Lanes: 1, P. aeruginosa M1134; 2, P. aeruginosa M1209; 3, P. aeruginosa UC2924; 4, P. aeruginosa PC483;5, E. coli J53 M1134; 6, E. coli J53 M1209; 7, E. coli J53 UC2924; 8, E. coli J53 PC483; 9, 100 bp Ladder
Screening for the Carbapenemase Production
By disc diffusion, all the P. aeruginosa isolates with reduced susceptibilities to meropenem and imipenem (the diameters of the zones of inhibition were ≤ 15mm) were screened for the production of carbapenemase. The phenotypic detection of the carbapenemase production was performed by the modified Hodge test by using a meropenem disc (10 μg) as per the CLSI guidelines [6]. The screening for the metallo-beta-lactamase production was performed by the combined – disc test, by using two imipenem discs (10 μg), one containing 10 μl of 0.1 M (292 μg) anhydrous EDTA (Sigma Chemicals, St. Louis, MO), which were placed 25 mm apart on a Mueller Hinton agar plate. An increase in the zone diameter of >4 mm around the imipenem-EDTA disc as compared to that around the imipenem disc alone was considered as positive for the metallo-β-lactamase production. The MBL production of the isolates was detected by the MBL (IP/IPI) E-test method (AB Biodisk, Solna, Sweden) as per the manufacturer’s instructions.
Molecular Detection of The MBL Genes
DNA was extracted by using the spin column method (QIAGEN; GmbH, Hilden, Germany) as per the manufacturer’s instructions. A multiplex PCR assay was performed to detect five families with the acquired MBL genes (blaIMP, blaVIM, blaSPM, blaGIM, blaSIM) in a single reaction. Multiplex PCR for the blaOXA-23 and the blaOXA-24 genes and simple PCR for blaKPC were carried out on the isolates by using the Gene Amp 9700 PCR System (Applied Biosystems, Singapore). The primers and the cycling conditions for the PCR were as has been described earlier [7–9]. PCR for the detection of blaNDM-1 was carried out by using primers, as has been reported previously [10]. Briefly, the program consisted of an initial denaturation step at 95°C for 5 min; followed by 35 cycles of denaturation at 95°C for 30s, primer annealing at 49°C for 30s, and extension at 72°C for 30s; followed by a final extension for 10 min at 72°C, which resulted in a 813 bp product. The amplicons were purified by using the QIAquick PCR purification kit (QIAGEN; GmbH, Hilden, Germany) and they were sequenced by using the ABI 3730XL capillary sequencer (Applied Biosystems, Foster City, CA, USA). The sequencing results were analyzed by using the Blast software which is there on the website of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov), which showed the presence of the blaNDM-1 gene. The plasmid DNA was extracted by using the QIAGEN plasmid Maxi kit as per the manufacturer’s instructions and the blaNDM-1 gene was detected in the plasmid by PCR analysis, as has been mentioned above.
Transfer of the resistance determinants and the plasmid analysis
Transfer of the resistance genes by conjugation was assayed by doing mating experiments in Luria–Bertani broth by using the P. aeruginosa isolates as the donors and an azide-resistant E. coli J53 strain as the recipient strain. The transconjugants were selected on Luria–Bertani agar, with the selection being based on the growth on the agar, in the presence of ceftazidime (30 mg/L) and sodium azide (100 mg/L). The plasmid DNA was extracted from the E. coli J53 recipient strain by using the QIAGEN plasmid Maxi kit (QIAGEN GmbH, Hilden, Germany) as per the manufacturer’s instructions and the blaNDM-1 gene was detected by PCR, as has been mentioned above. The phenotypic drug resistance assay which was done on the E. coli J53 transconjugants revealed resistance to the same carbapenems as the donor strain.
Molecular typing of the strains
Repetitive element based PCR (REP – PCR) and Enterobacterial Repetitive Intergenic Consensus (ERIC – PCR) assays were performed as have been described, to rapidly characterize the blaNDM-1 positive P. aeruginosa strains which were recovered from the patients [11].
Results
Out of the 200 clinical isolates of P. aeruginosa, 40 were found to be carbapenem resistant. The screening for MBL production was carried out on these 40 isolates, and 20 of them found to be positive. Among these 20 isolates, sixteen were found to be positive for blaVIM and four were found to be positive for blaNDM-1. The PCR results were validated by sequencing and the sequence of the blaNDM-1 gene showed a 100% identity with the previously reported genes [Table/Fig- 1]. The MIC values of these four isolates have been shown in [Table/Fig- 2].
Antibiotic susceptibilities of blaNDM-1-positive Pseudomonas aeruginosa (mg/L)
| Antibiotics | PS M1134 | PS M1209 | PS UC2924 | PS PC483 |
|---|
| IPM | 32 | 32 | 32 | 32 |
| MEM | 32 | 32 | 32 | 32 |
| AMK | 128 | 128 | 128 | 128 |
| TIC | 128 | 128 | 128 | 128 |
| GEN | 32 | 32 | 32 | 32 |
| TOB | 32 | 32 | 32 | 32 |
| CIP | 8 | 8 | 8 | 8 |
| LVX | 16 | 16 | 16 | 16 |
| MXF | 16 | 16 | 16 | 16 |
| TGC | 16 | 16 | 16 | 16 |
| SXT | 320 | 320 | 320 | 320 |
| SAM | 64 | 64 | 64 | 64 |
| TZP | 128 | 128 | 128 | 128 |
| FEP | 128 | 128 | 128 | 128 |
| SFP | 128 | 128 | 128 | 128 |
| CRO | 128 | 128 | 128 | 128 |
| CAZ | 128 | 128 | 128 | 128 |
| TET | 32 | 32 | 32 | 32 |
| CST | <0.5 | <0.5 | <0.5 | <0.5 |
IPM, imipenem; MEM, meropenem; TIC,ticarcillin ;AMK, amikacin; GEN, gentamicin ;TOB, tobramycin; CIP,ciprofloxacin; LVX,levofloxacin; MXF,moxifloxacin; TGC,tigecycline; SXT,trimethoprim/sulfamethoxazole; SAM, ampicillin/sulbactam; TZP,piperacillin/tazobactam; SFP cefoperazone/sulbactam; FEP, cefepime; TET,tetracycline; CAZ, ceftazidime; CRO,ceftriaxone; CST, colistin
Isolate 1 and Isolate 2 were from central venous catheter tip cultures. Isolate 1 was recovered from a 66 year old male patient with a necrotizing soft tissue infection (NSTI) in the left lower limb, with septicaemia. Isolate 2 was recovered from a 63 year old male patient who had developed septicaemia after undergoing cholecystectomy. Both the patients were successfully treated with colistin.
Isolate 3 was recovered from the urine sample of a 56-year old male who had pyelonephritis with pyuria. He had been treated with meropenem during his previous hospital admissions. He was successfully treated with colistin and amikacin.
Isolate 4 was recovered from the pus sample of a male patient, who was a case of a road traffic accident with a compound comminuted fracture of the tibia (left), and had developed a surgical site infection.
NDM-1 was also detected by PCR amongst the transconjugant E. coli J 53 strains, which indicated that the blaNDM-1 gene was located on a conjugative plasmid. Gel electrophoresis of the plasmid DNA which was extracted from the donor and the recipient strains demonstrated that the blaNDM-1 gene was present on a 50 kb transferable plasmid. The clonality of P. aeruginosa was confirmed by REP – PCR and ERIC – PCR and they confirmed that the isolate, 1, 2 and 4 were clonally similar and that isolate 3 was clonally different, as per the banding pattern.
Discussion
With the widespread use of the carbapenems, the emergence of NDM-1 was first detected in a Klebsiella pneumoniae isolate in 2008, in a Swedish patient of Indian origin; it has since been reported in increasing numbers of infections in patients from all over world. Nagarajan et al., have reported a high prevalence of blaNDM-1 in carbapenem resistant K. pneumoniae (75% of the isolates) and E. coli (66%), in a study which was done in the southern part of India [12]. In a similar study which was done at a tertiary care hospital in the northeastern region of India, all the isolates of K. pneumoniae which were carbapenem resistant were found to harbour the blaNDM-1 gene [13]. We have also reported the first instance of the detection of the blaNDM-1 gene in a clinical isolate of Raoultella ornitholytica [14]. Previous studies have revealed that the genes which encoded NDM-1 were mostly located on plasmids and these studies had focused on the Enterobacteriaceae [15]. In the present study, we confirmed that the blaNDM-1 genes of each of the four NDM-positive P. aeruginosa isolates were located on plasmids which were 50 kb in size. As all the four isolates of P. aeruginosa which carried blaNDM-1 were obtained from hospital acquired infection cases, the ability of blaNDM-1 to be transferred from P. aeruginosa to other Enterobacteriaceae and vice versa assumed significance, since this could result in the spread of multidrug resistant pathogens which caused untreatable infections in the hospital environment at an unprecedented rate.
Conclusion
P. aeruginosa has been a pathogen which is known to be notorious for the antibiotic resistance in hospital acquired infections, it causes severe infections in cases of burns, respiratory tract infections in patients who are on ventilators, urinary tract infections in catheterised patients and soft tissue infections which include surgical site infections. The carbapenems are the most frequently used antibiotics for the treatment of these infections. Resistance to the carbapenems requires the use of drugs like Colistin, which are more toxic to the patients. The accurate identification and the reporting of the NDM-1 producing P. aeruginosa will aid in the infection control management, for preventing its spread, especially when the isolates are multidrug resistant. The NDM-1 gene has been reported in Enterobacteriaceae. This is the first report on the detection of MBL NDM-1 in the clinical isolates of Pseudomonas aeruginosa in India.
The nucleotide sequence data of the blaNDM-1 gene from the four isolates of P. aeruginosa which have been reported in the present study have been assigned the GenBank nucleotide numbers, JX680682, JX680683, JX680684 and JX680685, respectively.
IPM, imipenem; MEM, meropenem; TIC,ticarcillin ;AMK, amikacin; GEN, gentamicin ;TOB, tobramycin; CIP,ciprofloxacin; LVX,levofloxacin; MXF,moxifloxacin; TGC,tigecycline; SXT,trimethoprim/sulfamethoxazole; SAM, ampicillin/sulbactam; TZP,piperacillin/tazobactam; SFP cefoperazone/sulbactam; FEP, cefepime; TET,tetracycline; CAZ, ceftazidime; CRO,ceftriaxone; CST, colistin