Dermatophytes are the most common, superficial, filamentous fungi that cause skin, hair and nail infections. They belong to three anamorphic genera which are classified, based on the conidial morphology and the accessory structures- the Trichophyton, Microsporum and the Epidermophyton [1]. They have the ability to utilize the keratin (keratinophilic) and to destroy the keratinized tissues (keratinolytic) of the host [2]. They usually colonize the nonliving, cornified layer of the epidermis, as they are unable to penetrate the deeper tissues of an immunocompetent host. The infection which is caused by these fungi is termed as dermatophytosis and it is commonly referred to as ringworm or tinea. Poor hygienic conditions, over population and a highly humid weather are the causative factors of dermatophytosis.
The standard phenotypic identification of the dermatophytes depends on the appropriate culture media, followed by the macroscopic examination of the colony characteristics (colour, texture, pigmentation on the obverse and the reverse sides, topography and the rate of growth) and the microscopic morphology (size and shape of the macro and microconidia, spiral hyphae, arthroconidia, nodular organ, chlamydospores, favic chandeliers, etc). The further identification includes the urease production, pigment production on corn meal agar and the in-vitro hair perforation test.
Though culture based identification is a gold standard method, it is time-consuming, as it requires 14 - 21 days for the growth of the organism and to observe the typical features in identification of the dermatophyte species directly from the clinical specimens. Although a dermatophyte infection is not an emergency, identification of the dermatophyte species is essential, to rule out the lesions which simulate dermatophytosis and hence, start the appropriate treatment at the earliest.
In the past few years, molecular typing methods have proven to be useful for a rapid detection and identification of the dermatophyte species. In fact, a genotypic identification is considered to be more stable and precise than the phenotypic methods. Recently, a number of genetic advances in dermatophytes have been reported, which include - targeted gene inactivation, gene silencing and transcriptional profiling methods [6]. Whole genome sequencing [7] was also developed to study the future outbreaks on the biology, virulence, pathogenicity and the host specificity of the clinically important dermatophytes. Molecular typing is essential for the identification of the fungal isolates upto the genus, species and the strain levels for epidemiological purposes. Genotypic methods such as arbitrarily primed PCR (AP-PCR) [8], random amplified polymorphic DNA (RAPD) [9,10], repetitive sequence PCR (rep-PCR) [11], restriction analysis of the mitochondrial DNA [12,13], semi-nested PCR [14], nested PCR [15], multiplex PCR [16] and single-strand conformation polymorphism (SSCP) analysis [17], are the available techniques for the identification of dermatophytes. However, few methods have reported a low sensitivity and specificity in the identification of the dermatophyte species. Therefore, the present study was performed to compare the two PCR based typing methods – the pan fungal primer targeting of the Internal Transcribed Spacer (ITS) region and dermatophyte specific primer targeting of the 18S ribosomal DNA (rDNA) region. Only those strains which were positive on using the dermatophyte specific primer were subsequently, digested with the Mva I, Hae III and Dde I restriction enzymes separately for an accurate identification of the dermatophyte species, as well as the strains.
MATERIALS AND METHODS
Clinical Specimens
One hundred and thirty eight specimens (129 skin scrapings and 9 nail clippings) of clinically suspected dermatophytosis, who attended the Dermatology Outpatient Department of a tertiary care centre, were collected between January – December 2010 and they were processed by direct microscopy and culture. Out of the 138 clinical specimens, 69 were taken up for molecular studies (as the specimens which were collected from all the cases were not adequate), which comprised of 66 skin scrapings and 3 nail clippings for the genotypic identification of the dermatophyte species and strains. An ethical approval was obtained from the institutional review board for performing the study (IEC-NI/09/DEC/13/40).
Phenotypic Methods
The clinical specimens were subjected to 10% KOH mount and they were inoculated onto Sabouraud’s Dextrose Agar (SDA) that contained gentamicin and cycloheximide and onto the Dermatophyte Test Medium (DTM), all in duplicates and the plates were incubated at 250C and 370C. The dermatophytes were identified, based on their colony morphology, their microscopic examination in the Lactophenol Cotton Blue (LPCB) preparations and on the slide culture techniques. The further identification of the dermatophyte species was based on the urease production, pigment production on corn meal agar and on the in-vitro hair perforation test.
Optimization of PCR Directly from the Skin and Nail Specimens
Extraction of the Genomic DNA
The genomic DNA was directly extracted from the clinical specimens by using the Omni PrepTM kit for fungus (G Biosciences, USA) according to the manufacturer’s instructions. In brief, the clinical specimens (skin scrapings and nail clippings) were inoculated in 500μl of genomic lysis buffer along with 30 μl of molecular grinding resin and they were ground by using a mortar and pestle. 5 μl of Proteinase K was added and the samples were incubated at 600C for 2 hours. In case of the nail specimens, 15 μl of Proteinase K was added and they were incubated overnight at 600C. After adding 200 μl of chloroform, the samples were spun at 14000 rpm for 10 min. The supernatant was taken, 50 μl of DNA stripping solution was added and the vials were incubated at 600C for 10 min. 100 μl of the precipitation solution was added and the vials were spun at 14000 rpm for 5 min. To the supernatant, ice cold isopropyl alcohol was added to precipitate the DNA. Finally, the DNA was washed with 70% ethanol and air dried and it was re-suspended with 100 μl of Tris EDTA buffer. It was stored at -200C for further use.
Diagnostic Sensitivity: A logarithmic ten-fold serial dilution was done with the ATCC 28188 strain of T. rubrum, which ranged from 10-1 to 10-10 and it was analyzed to estimate the diagnostic sensitivity of the PCR assay.
Diagnostic Specificity: Two reference strains, T. mentagrophytes ATCC 9533 and T. rubrum ATCC 28188 (HiMedia, India) were used to ascertain the specificity of the dermatophyte specific primer. Laboratory isolates of Candida albicans, Candida parapsilosis and Fusarium species were also used to test the specificity of the pan fungal primers.
Extraction of the Bacterial DNA: Bacterial organisms (Staphylococcus aureus ATCC 25923 and Pseudomonas aeruginosa ATCC 27853) that had the ability to produce common skin infections were used to define the specificity of the fungal primers. The bacterial DNA was extracted by the boiling method.
Pan Fungal Primers which targeted the ITS Region
Uniplex PCR was performed by using pan fungal primers ITS 1 (5’- TCC GTA GGT GAA CCT GCG G- 3’) and ITS 4 (5’-TCC TCC GCT TAT TGA TAT GC - 3’). The primers and the PCR reagents were purchased from Bangalore Genei Pvt. Ltd, Bangalore, India. The PCR was carried out by using the Eppendorf Master Cycler Gradient (Model 5331). The PCR reaction mixture constituted of 200 μM concentration of each dNTP, 25 pmol of each primer, 1 U of Taq polymerase, 5 μl of Taq buffer and 10 μl of template DNA. Sterile nuclease free water was added to make up the final volume to 50 μl. The PCR amplification conditions consisted of an initial denaturation at 950C for 5 min, followed by 35 cycles at 950C for 30s, 550C for 1 min and 720C for 1 min and a final extension at 720C for 5 min.
Dermatophyte Specific Primers which targeted the 18S rDNA Region
Uniplex PCR was carried out by using the published dermatophyte specific primers DH1L (5’ – TGC ACT GGT CCG GCT GGG – 3’) and DH1R (5’ – CGG CGG TCC TAG AAA CCA AC – 3’) [18]. The amplification was performed in a reaction mixture which contained 200 μM concentration of each dNTP, 25 pmol of each primer, 1 U of Taq polymerase and 10 μl of template DNA. This mixture was made up to 50 μl by using sterile nuclease free water. The PCR thermal conditions comprised of an initial denaturation at 950C for 3 min, followed by 35 cycles at 950C for 1 min, 580C for 1 min and 720C for 40 s and a final extension at 720C for 5 min.
Parallel to each PCR assay using the pan fungal primer and the dermatophyte specific primer, inhibitory and cross – contamination controls were performed, with either 2.5 μl of positive control or 5 μl of water respectively.
Detection of the Amplified Products: The PCR amplified products were electrophoretically separated in a 2% agarose gel in 1X Tris acetate – EDTA buffer and they were visualized by using ethidium bromide, under a UV trans-illuminator (Bio-Rad Laboratories India Pvt. Ltd).
PCR – RFLP Analysis
The specimens which are positive on the use of the dermatophyte specific primer, were taken up for the RFLP analysis. The non-dermatophytes, which are positive only on ITS PCR and not on amplification of the dermatophyte specific primer, were not included in the present study.
Both the PCR amplified products were digested separately by using the Hae III (Bangalore Genei Pvt. Ltd, Bangalore, India), Mva I and the Dde I (Fermentas Inc. USA) restriction enzymes and they were incubated at 370C for 2 hours. The digested products were electrophoresed in a 2 % agarose gel, stained with ethidium bromide and observed under a UV trans-illuminator.
DNA sequencing
PCR based DNA sequencing targeting the ITS region was performed at the Vision Research Foundation (VRF) Referral Laboratory (A unit of Sankara Nethralaya, Chennai, India) [19]. Due to various constraints, only few of the representative isolates were sent for further confirmation. The PCR product was sequenced by the dideoxy chain termination method by using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, USA).
STATISTICAL ANALYSIS
The study data was analyzed by using the statistical software program: Open Epi (Open Source Epidemiologic statistics for Public health; Version 2.3.1). The PCR and direct microscopy were compared against the gold standard culture method. The sensitivity, specificity, positive predictive value, negative predictive value and the 95% Confidence Interval (CI) were calculated.
RESULTS
Of the 138 specimens (129 skin scrapings and 9 nail clippings), 81 were positive for dermatophytosis. Fifty-one were males and 87 were females, and their age ranged between 6 – 80 years. A majority of the patients were between 30 – 40 years. The patients had scaly and papulo erythematous lesions on their faces, arms, shoulders, armpits, lower abdomen, buttocks, thighs and groin. In our study, tinea corporis was found in 80 (57.97%) cases with dermatophytosis, followed by tinea cruris, tinea unguium, tinea faciei and tinea pedis in 34 (24.63%), 9 (6.52%), 3 (2.17%) and 1 (0.72%) cases respectively. Eleven patients (7.97%) had mixed infections of tinea corporis and tinea cruris.
Basic Laboratory Identification
Among the 129 skin scrapings, direct microscopy and culture showed the presence of dermatophytes in 51.16% (66/129) and 58.13% (75/129) cases respectively. The predominant dermatophyte isolated was T. rubrum (43), followed by T. mentagrophytes (23) and Epidermophyton floccosum (9). Of the 54 negative cases of dermatophytosis, six cases were positive for the Candida species in culture. Of the nine nail clippings, only one was positive for dermatophytosis, both by direct microscopy, culture and was identified as T. rubrum. Two specimens were positive for non-dermatophytic molds (Fusarium spp.) and two cases were positive for the Candida species in culture.
Molecular Typing
PCR sensitivity: The diagnostic sensitivity of the PCR assay on the use of the pan fungal primer and the dermatophyte specific primer was 10 picograms of the T. rubrum ATCC 28188 strain.
PCR specificity: The dermatophyte specific primer which was used in this study selectively amplified the standard strains of the dermatophytes which were tested and it did not amplify the non-dermatophytic fungi. The pan fungal primer broadly amplified the standard fungal isolates, including the dermatophytes. The amplification product was not detected by using the dermatophyte specific primer and the pan fungal primer with the DNA extracted from the ATCC strain of S. aureus and P. aeruginosa.
Genotypic Identification from the Skin and Nail Specimens
Of the 66 skin scrapings, 54.54% (36/66) cases were positive by direct microscopy, 63.63% (42/66) cases were positive on culture and forty-seven (71.21%) cases were positive on PCR for dermatophytosis, of which five cases were positive on PCR alone. Dermatophytes were identified by PCR using the pan fungal primer which targeted the ITS region, resulting in an amplified product size of 690 bp for the T. mentagrophytes and the T. rubrum strains, whereas E. floccosum showed a larger fragment size of 740 bp [Table/Fig-1]. Subsequently, the dermatophyte specific primer which targeted the 18S rDNA region was used, which resulted in an approximately 180 bp for the dermatophytes [Table/Fig-2]. The clinical and the statistical data are shown in [Table/Fig-3]. Of the 19 PCR negative cases of dermatophytosis, two cases were identified as Candida species by PCR and they were also positive on culture. The ITS PCR produced an amplified product size of 595 bp, but the PCR showed negative results on the use of the dermatophyte specific primer.
Amplification using pan fungal primer on clinical specimens. Lane 1 … 17: 100 bp DNA ladder; Lane 2: Negative control (no template DNA); Lane 3: S. aureus; Lane 4: P. aeruginosa; Lane 5: C. albicans - 595 bp; Lane 6: C. parapsilosis - 595 bp; Lane 7: Patient no. 70 - Candida spp; Lane 8: Fusarium species - 550 bp; Lane 9: Patient no. 43 – Fusarium spp; Lane 10: T. rubrum ATCC - 690 bp; Lane 11: T. mentagrophytes ATCC - 690 bp; Lane 12 – 15: Patient samples – positive; Lane 16: Patient sample- negative.
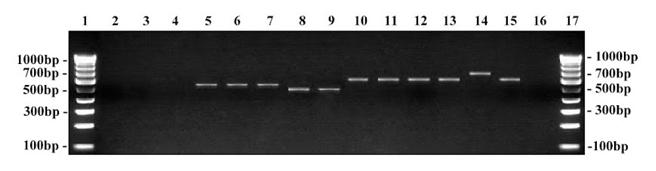
Amplification using dermatophyte specific primer on clinical specimens. Lane 1 … 17: 100 bp DNA ruler; Lane 2: Negative control (no template DNA); Lane 3: S. aureus; Lane 4: P. aeruginosa; Lane 5: C. albicans; Lane 6: C. parapsilosis; Lane 7: Patient no. 70 – negative; Lane 8: Fusarium species; Lane 9: Patient no. 43 – negative; Lane 10: T. rubrum ATCC - 180 bp; Lane 11: T. mentagrophytes ATCC - 180 bp; Lane 12 – 15: Patient samples – positive; Lane 16: Patient sample – negative.
Clinical and statistical analysis of the diagnostic methods of skin scrapings
| Comparative Analysis of Smear vs Culture for 129 Skin Scrapings |
|---|
| Culture Positive (75) | Culture Negative (54) | Sensitivity (CI) | Specificity (CI) | Positive Predictive value (CI) | Negative Predictive Value (CI) |
|---|
| Smear positive (66) | 59 | 7 | 78.67 % (68.12-86.42) | 87.04 % (75.58-93.58) | 89.39 % (79.69-94.77) | 74.6 % (62.66-83.72) |
| Smear negative (63) | 16 | 47 |
| Comparative Analysis of Smear, Culture and PCR for Diagnosis of Dermatophytes (66 skin scrapings) |
| Culture Positive (42) | Culture Negative (24) | Sensitivity (CI) | Specificity (CI) | Positive Predictive value (CI) | Negative Predictive Value (CI) |
| Smear positive (36) | 33 | 3 | 78.57% (64.06-88.29) | 87.5% (69-95.66) | 91.67% (78.17-97.13) | 70% (52.12-83.34) |
| Smear negative (30) | 9 | 21 |
| PCR positive (47) | 42 | 5 | 100% (91.62-100) | 79.17% (59.53-90.76) | 89.36% (77.41-95.37) | 100% (83.18-100) |
| PCR negative (19) | 0 | 19 |
Among the three nail samples, one specimen was positive on PCR for the dermatophytes, which was also positive on the direct microscopy and culture. Of the two cases which were negative on PCR, on the use of the dermatophyte specific primer, one was positive for the Candida species and the other one was positive for the Fusarium species in culture, which was also positive by ITS PCR, with amplified product sizes of 595 bp and 550 bp respectively.
The PCR was performed with clinical specimens at different periods and the band patterns which were obtained were consistent. They were found to be highly reproducible.
PCR-RFLP and DNA Sequencing
The dermatophyte species and strains were identified by the PCR-RFLP method. The band profiles obtained by using the pan fungal primer which targeted the ITS region on the Mva I, Hae III and the Dde I enzymes are depicted in [Table/Fig-4, 5 & 6]. Similarly, the RFLP profiles obtained by using the dermatophyte specific primer which targeted the 18S rDNA region on the three restriction enzymes are depicted in [Table/Fig-7, 8 & 9].
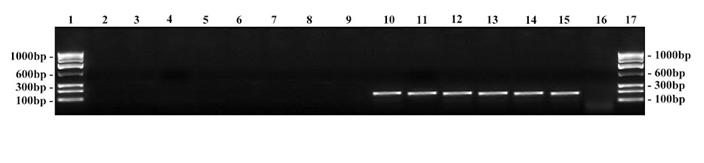
RFLP on ITS amplicons using Mva I restriction enzyme. Lane 1: 100 bp DNA ladder; Lane 2: Undigested product of T. rubrum ATCC - 690 bp; Lane 3 – 10: Digested products; Lane 3: T. mentagrophytes ATCC - 250, 180, 160, 120 bp; Lane 4: Clinical strain - T. mentagrophytes; Lane 5: Clinical strain - T. interdigitale; Lane 6: T. rubrum ATCC - 400, 180, 120 bp; Lane 7: Clinical strain - T. rubrum; Lane 8: Clinical strain - T. rubrum var. raubitschekii; Lane 9 & 10: Clinical strain - E. floccosum - 400, 250, 180 bp.
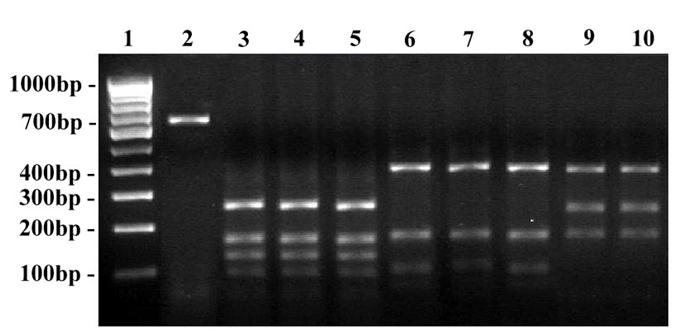
RFLP on ITS amplicons using Hae III restriction enzyme. Lane 1: Undigested product of T. rubrum ATCC - 690 bp; Lane 2 - 9: Digested products; Lane 2: T. mentagrophytes ATCC - 400, 90 bp; Lane 3: Clinical strain - T. mentagrophytes; Lane 4: Clinical strain - T. interdigitale; Lane 5: T. rubrum ATCC - 320, 100 bp; Lane 6: Clinical strain - T. rubrum; Lane 7: Clinical strain - T. rubrum var. raubitschekii; Lane 8 … 9: Clinical strain - E. floccosum - 400, 90 bp; Lane 10: 100 bp DNA ladder.
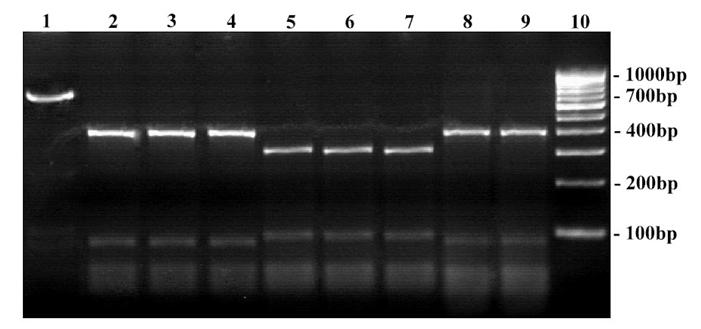
RFLP on ITS amplicons using Dde I restriction enzyme. Lane 1: Undigested product of T. rubrum ATCC - 690 bp; Lane 2 – 9: Digested products; Lane 2: T. mentagrophytes ATCC - 400, 290 bp; Lane 3: Clinical strain - T. mentagrophytes; Lane 4: Clinical strain - T. interdigitale; Lane 5: T. rubrum ATCC - 300, 290, 100 bp; Lane 6: Clinical strain - T. rubrum; Lane 7: Clinical strain - T. rubrum var. raubitschekii; Lane 8 … 9: Clinical strain - E. floccosum - 500, 290 bp; Lane 10: 100 bp DNA ladder.
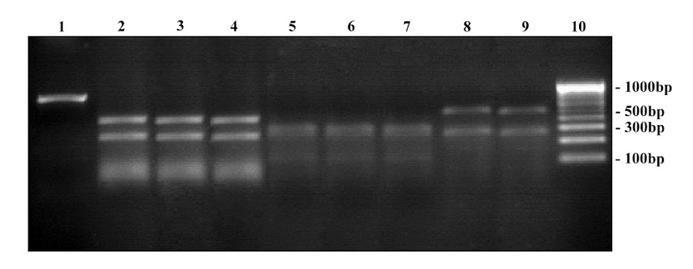
RFLP on 18S rDNA amplicons using Mva I restriction enzyme. Lane 1: Undigested product of T. rubrum ATCC - 180 bp; Lane 2 – 9: Digested products; Lane 2: T. mentagrophytes ATCC - 100, 80 bp; Lane 3: Clinical strain - T. mentagrophytes; Lane 4: Clinical strain - T. interdigitale; Lane 5: T. rubrum ATCC - 100, 90 bp; Lane 6: Clinical strain - T. rubrum; Lane 7: Clinical strain - T. rubrum var. raubitschekii; Lane 8 … 9: Clinical strain - E. floccosum - 100, 90 bp; Lane 10: Hinf – I digest of ΦX174 bacteriophage DNA ladder.
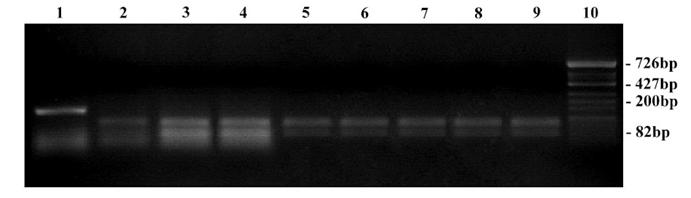
RFLP on 18S rDNA amplicons using Hae III restriction enzyme. Lane 1: Undigested product of T. rubrum ATCC - 180 bp; Lane 2 – 9: Digested products; Lane 2: T. mentagrophytes ATCC - 100, 90 bp; Lane 3: Clinical strain - T. mentagrophytes; Lane 4: Clinical strain - T. interdigitale; Lane 5: T. rubrum TCC - 100, 80 bp; Lane 6: Clinical strain - T. rubrum; Lane 7: Clinical strain - T. rubrum var. raubitschekii; Lane 8 & 9: Clinical strain - E. floccosum - 100, 90 bp; Lane 10: Hinf-I digest of ΦX174 bacteriophage DNA ladder.
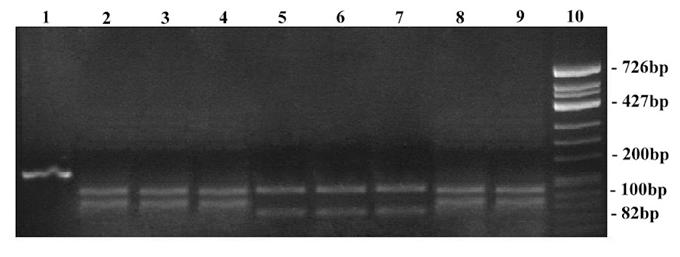
RFLP on 18S rDNA amplicons using Dde I restriction enzyme. Lane 1: Undigested product of T. rubrum ATCC - 180 bp; Lane 2 – 9: Digested products; Lane 2: T. mentagrophytes ATCC - 180 bp; Lane 3: Clinical strain - T. mentagrophytes; Lane 4: Clinical strain - T. interdigitale; Lane 5: T. rubrum ATCC - 180 bp; Lane 6: Clinical strain - T. rubrum; Lane 7: Clinical strain - T. rubrum var. raubitschekii; Lane 8 … 9: Clinical strain - E. floccosum - 180 bp; Lane 10: 100 bp DNA ruler
The query sequences were paired with those in the GenBank database by Blast analysis. Of the 47 T. rubrum isolates, ten were T. rubrum var. raubitschekii, which were identified phenotypically as urease positive and by ITS sequencing. The representative isolates displayed 99% and 100% identities of both T. rubrum var. raubitschekii ATCC 42631 and T. rubrum ATCC 28188. Of the 25 T. mentagrophytes isolates, three were confirmed as T. interdigitale, of which two displayed 99% similarity and one displayed 100% similarity of the T. interdigitale strains.
DISCUSSION
Superficial mycotic infections account for more than 20-25% of the infections in the world’s population and they are predominantly caused by dermatophytes [3]. The most common dermatophyte species which were isolated in our study were T. rubrum and T. mentagrophytes, which are predominant worldwide, but less frequently reported in Africa [3]. E. floccosum was less frequently isolated in the present study.
Molecular techniques can be used as an epidemiological tool for the detection of dermatophytes. In the identification of dermatophytes, molecular methods have an edge over the conventional procedures, which are either slow in diagnosis or lack enough specificity. They may not detect all the true positives. The standard laboratory identification can easily identify dermatophytes upto the genus level but for the identification of the species and strains, these fungi, on sub-culture, show different characteristic colony morphology. Therefore, an accurate identification of dermatophytes at the species or the strain levels can be done best by using molecular methods for epidemiological surveys.
In the present study, we compared 2 PCR based typing methods for the direct identification of dermatophytosis from skin and nail specimens. Among the 66 skin scrapings and 3 nail clippings, the dermatophyte specific primer that targeted the 18S rDNA region specifically, amplified the dermatophyte DNA and it did not amplify the other fungi. Therefore, the dermatophyte specific primer is specific and it is accurate in the direct identification of dermatophytosis from clinical material. Whereas, the pan fungal primer which targeted the ITS region, amplified all the fungal DNA, including the dermatophytes. Therefore, in the identification of dermatophytosis, the application of the pan fungal primer on the direct clinical specimens may not be specific.
Furthermore, the dermatophytes that were positive on culture, showed positivity by PCR on the use of the dermatophyte specific primer and they were highly reproducible. Therefore, it can be concluded that PCR detected all the true positives with a 100% correlation.
In the present study, on opting culture as a gold standard method, the sensitivity of the smear and PCR were 78.57% and 100% respectively. Therefore, the diagnostic sensitivity of PCR is high enough to pick up the dermatophytosis from the skin and nail specimens. In our study, the diagnostic specificity of PCR was 79.17%. Of the 47 PCR positive cases, five cases were positive on PCR alone, and these patients responded well to the antifungal treatments. The reason for the culture negativity in the PCR positive cases could be the inadequate fungal load in the specimen and therefore, the culture method had probably failed to enrich the growth of dermatophytes.
The RFLP product using the dermatophyte specific primer on the Mva I and Hae III enzymes showed an identical band size consistently and with the Dde I enzyme, it showed no recognition site on the dermatophytes which were tested. Therefore, there was no difference in the band patterns. Eventually, using the dermatophyte specific primer, followed by RFLP, produced similar band profiles and this made the identification and the speciation of the dermatophytes difficult. In case of the pan fungal primer, using the Hae III enzyme based RFLP speciation, produced similar band patterns and therefore, the Hae III enzyme may not be suitable for the identification of the dermatophyte species. As was described previously, using the Mva I [20] and the Dde I restriction enzymes produced unique band profiles consistently and it was reproducible. Therefore, the application of the Mva I and the Dde I enzymes by using the ITS amplicons helped in the easy identification of the dermatophyte species. However, by using the dermatophyte specific primer and the pan fungal primer with these three restriction enzymes, it was not possible to detect any strain variations among the T. rubrum and the T. mentagrophytes strains. Therefore, in the identification of the strain variations by using RFLP analysis, the recognition site for dermatophytes was not found to be located in the ITS and the 18S rDNA regions. As was described in earlier studies, the strain variations can be identified by targeting the ribosomal DNA of the Non-Transcribed Spacer (NTS) region [21].
DNA sequencing confirmed the isolates as T. rubrum var. raubitschekii, which were identified phenotypically as urease positive. Since T. rubrum var. raubitschekii possessed minor morphological and physiological features, it is currently being considered as a synonym of T. rubrum. The previous report on T. interdigitale from India was made in 1996 [22] and the present report is the second one from India.
In the present study, among the non-dermatophytes (0.07%), the Candida (8) and Fusarium (2) species were the isolates that were grown from skin and nails. In general, onychomycosis is a common fungal infection, which accounts for upto 50% of the fingernail and toenail infections [23]. Weinberg et al., (2003) reported that the non-dermatophytes constituted approximately 10% of the causative agents of onychomycosis [24], whereas El Batawi et al., (2007) reported about 68.75% were non-dermatophytes and 0.1% were dermatophytes that caused onychomycosis [25].
To conclude, the dermatophyte specific primer based PCR which targets the 18S rDNA is useful in the direct identification of dermatophytosis from clinical specimens and it can be applied in the routine diagnostics wherever the laboratory facilities are adequate. The application of the Mva I and the Dde I restriction enzymes by using the ITS amplicons was equally good, stable and reproducible in the identification of the dermatophyte species. The PCR-RFLP method, on using the dermatophyte specific primer and the pan fungal primer with Mva I, Hae III and Dde I, showed no strain differentiation among the T. rubrum and the T. mentagrophytes isolates. Since direct microscopy and culture have limitations, performing a direct PCR on the clinical specimens can augment the diagnosis of more dermatophyte cases. However, species identification by PCR may not have a direct impact on the clinical treatment.