The Extended Spectrum Beta Lactamases {ESBLs} are enzymes which are produced by gram negative bacilli, that mediate a resistance to the penicillins, cephalosporins and the monobactams and are commonly recognized in Enterobacteriaceae. Gram negative bacteria are the major pathogens in hospitalized patients with weakened host defenses, due to malignancy, malnutrition, debilitation, trauma, a prolonged antibiotic use and surgery. The most frequently bacteria are E. coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Enterobacter and Acinetobacter baumanii. Once these organisms are established as the source of the infection, they are aggressively treated with antimicrobial drugs, which are thought to be the greatest contribution of the present century to therapeutics.
The first plasmid mediated beta lactamase in gram negative bacteria, TEM-1, was described in the early 1960s [1]. Being plasmid and transposon mediated, it quickly spread to various species of bacteria. SHV-1 was the next plasmid mediated beta–lactamase which was discovered [1]. Although most of the ESBLs are mutants of the TEM and the SHV enzymes, the CTX-M type beta lactamases have become more important. The CTX-M type of enzyme constitutes a distinct lineage of the molecular class A β lactamases, which are a rapidly growing group. Rather than arising by mutation, they represent examples of plasmid acquisition of the βlactamases genes which are found on the chromosomes of the Kluyvera species. CTX-M was 1st detected in E.coli strains in Germany (1988) and presently, there are more than 65 allelic variants which are known [2,3].
The detection of the common ESBL genes such as TEM, SHV and CTX-M by molecular methods in the ESBL producing bacteria and their patterns of antimicrobial resistance can provide useful information about their epidemiology and can aid a rational antimicrobial therapy.
The present study was therefore designed to investigate the prevalence of the ESBL producing gram negative organisms in the family, Enterobacteriaceae by phenotypic methods and to characterize the ESBL types which were prevalent in our hospital, by multiplex PCR.
MATERIAL METHODS
Samples
This study was done on 500 gram negative isolates which belonged to the family, Enterobacteriaceae in the Department of Microbiology in our Institute. Clinical isolates from urine (344), pus (109), blood (15), IV catheter tip/ central line tip (10), sputum (12) and body fluid (10) specimens were processed.
Antimicrobial Susceptibility Testing
The isolates were tested for their antimicrobial susceptibilities by the disc diffusion technique according to the CLSI guidelines [4]. The following antibiotic discs (drug concentrations in μg) were used: ampicillin (10), amikacin (30), ceftazidime (30), cefotaxime (30), ceftriaxone (30), cefepime (30), gentamicin (10), imipenem (10), ciprofloxacin (5), netilmicin (30), norfloxacin (10) and nitrofurantoin (300).
The detection of ESBLs
The ESBL detection was performed as was recommended by the CLSI confirmatory procedure, by using cefotaxime (30ug) and ceftazidime (30ug) discs alone and in combination with clavulanic acid discs [4]. E. coli (ATCC-25922) and K. pneumoniae (ATCC-700603) were used as the controls throughout the study.
Confirmation of the ESBLs was done by performing the E-test with a two sided strip which contained ceftazidime on one side and ceftazidime+clavulanic acid on the other side (which was procured from Biomerieux, India). An isolate was considered as an ESBL producer if the ratio of the MIC of the combination to that of ceftazidime alone was = 8 or if there was the development of a phantom zone [Table/Fig-1]. We used a modified method in which two strains were simultaneously swabbed on a single plate by dividing the plate into two halves, being separated by a 3mm wide uninoculated area where the E-test strip was placed.
Modified E-test (Tz/Ceftazidime, Tzl/Ceftazidime + Clavulanic Acid)
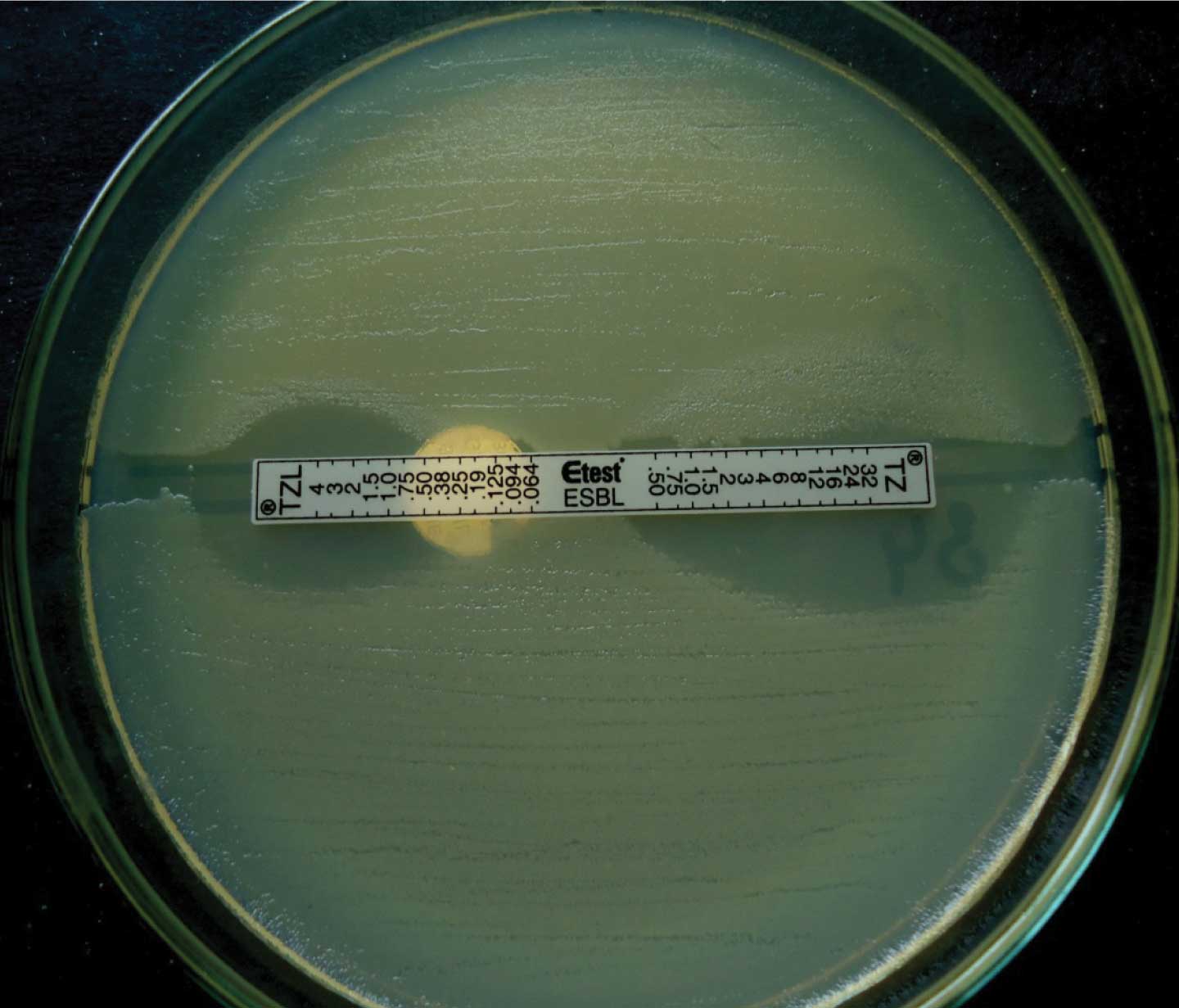
The Extended Spectrum β-lactamase Gene Identification
Multiplex PCR was carried out for the detection of the TEM, SHV and the CTX-M genes among 93 study isolates which were isolated from the ICU patient samples (which were determined as ESBL producers by phenotypic methods). PCR was used to characterize the type of ESBLs which were prevalent in our study, so that the strains that were found to be ESBL producers by both the CLSI confirmatory method and the E test method were genotyped. The DNA template preparation was performed as follows: 2 ml of an overnight broth culture (Luria Burtonni broth-Himedia) of the test organism was centrifuged at 8000rpm for 10 minutes. The supernatant was discarded and the pellet was mixed with 200 μl of sterile distilled water and it was vortexed. This was then boiled in a dry bath for 10 minutes and was immediately chilled by putting it on ice for 5 minutes. It was then centrifuged once again at 6000rpm and the supernatant which contained the nucleic acid material, was pipetted off and stored for genetic testing. The master mix for the PCR was prepared as follows: 2.5μL of PCR buffer, 2.5μL of 25mM MgCl2, 2.5μLof10mM dNTP mix, 0.3μL of Taq DNA Polymerase , 9.2μL of MilliQ water and 1μL of each of the forward and reverse primers. Finally, after dispensing 23μL of the master mix in the individual amplification tubes, 2μL of the extracted DNA was added in the corresponding tubes to make up the total volume to 25μL.The PCR primers which were used have been indicated in [Table/Fig-2] [5,6]. PCR was performed according to the following protocol: prehold for 2 min at 95°C, followed by 30 cycles, each consisting of a denaturation at 95°C for 45 sec, annealing at 62°C for 45 sec, extension at 72°C for 1 min and followed by a post extension hold at 72°C for 5min. The PCR products were analyzed by gel electrophoresis in a 1.5% agarose gel in 0.5% of TAE buffer. The gels were stained with 0.5μg/ml Ethidium bromide and the PCR products were visualized in UV light.
primer pairs used for amplification of the TEM, SHV and CTX-M ESBL types
| Target gene | Target gene |
|---|
| BlaTEM | TEM-F(5′-3′): GTA TCC GCT CAT GAG ACA ATA ACC CTG TEM-R (5′-3′): CCA ATG CTT AAT CAG TGA GGC ACC |
|
BlaSHV | SHV-F(5′-3′): CGC CTG TGT ATT ATC TCC CTG TTA GCC SHV-R (5′-3′): TTG CCA GTG CTC GAT CAG CG |
| BlaCTX-M | CTX-M (5′-3′): CGC TTT GCG ATG TGC AG ACC GCG ATA TCG TTG GT |
RESULTS
A total of 500 isolates of E.coli (n=351), Klebsiella pneumoniae (n=74), Klebsiella oxytoca (n=21), Proteus mirabilis (n=15), Proteus vulgaris (n=9) , Enterobacter spp (n=15) and Citrobacterspp (n=15) were recovered from different clinical specimens.
Of the total 500 isolates, 315/500 (63%) isolates were found to be resistant to at least three cephalosporins (3rd or 4th generation). The resistance to the piperacillin tazobactam combination was 9.97 % in E.coli and it was 12.24 % in the Klebsiella pneumoniae isolates. Among the non beta lactams, the maximum resistance was shown to norfloxacin and ciprofloxacin (79.67% and 91.66% in E.coli, 59.45% and 71.42% in Klebsiella pneumoniae). The E.coli and Klebsiella spp exhibited comparatively less resistance to amikacin and nitrofurantoin. (13.75% and 18.35% in E.coli, 42.59% and 34.14% in Klebsiella pneumoniae).
Among the total 500 isolates, ESBL production was noted in 45.8% (229 isolates) by the CLSI confirmatory test. The maximum ESBL production was seen among the isolates of Klebsiella pneumoniae (52.27%), followed by those of E.coli (46.43%). The ESBL production was quite high among the pus samples (51.37%), followed by urine samples (45.63%). By the ESBL E-test method, 220 isolates were found to have an MIC ratio of TZ/TZL, as more than 8 and so were confirmed as ESBL producers. The E test which was done by using ceftazidime and ceftazidime plus clavulanic acid showed that the 220/229 isolates which were detected by the confirmatory method were also found to be ESBL producers by the E test and that the difference was not statistically significant. So, the CLSI confirmatory method is as sensitive and as specific as the E test. The ESBL producing E.coli and K. pneumoniae were found to be quite sensitive to Imipenem (the resistance was only 2.9%) and a combination of piperacillin and tazobactam .
In the present study, the rate of the drug resistance [Table/Fig-3] was higher in the ESBL producers than in the non-ESBL producers. As for the beta-lactam antibiotics, the drug resistance was higher for other antibiotics also, namely, ciprofloxacin (83.22%), norfloxacin (84.95%) and gentamicin (57.74%). The statistical analysis showed a highly significant difference (a P value of less than 0.001) in the resistance for the non-beta lactam antibiotics like gentamicin, amikacin, norfloxacin, levofloxacin, ciprofloxacin and netilmicin among the ESBL producers and the non-ESBL producers.
Antimicrobial resistance pattern among ESBL producers and non-ESBL producers
| S. No. | Anti-microbial Agent | ESBL producers | Non ESBL producers | P value |
|---|
| 1. | Gentamycin | 57.74% | 30% | <0.001 |
| 2. | Amikacin | 34.19% | 16.31% | <0.001 |
| 4. | Norfloxacin | 84.95% | 50.72% | <0.001 |
| 5. | Levofloxacin | 54.19% | 31.05% | <0.001 |
| 6. | Ciprofloxacin | 83.22% | 60.52% | <0.001 |
| 7. | Netilmycin | 50% | 24.73% | <0.001 |
| 8. | Chloramphenicol | 56.73% | 30.76% | <0.05 |
Molecular Characterization
The 93 phenotypically confirmed ESBL producers were genotyped [Table/Fig-4]. The molecular characterization revealed that 44.08% isolates possessed the CTX-M genes alone, while the CTX-M and the SHV genes together were found in 20.43% isolates. 6.45% isolates which were genotyped, contained all the 3 genes, CTX-M, SHV and TEM. A majority of the E.coli (59.32%) isolates in the study carried the CTX-M genes and the Klebsiella isolates were found to be co producers of the ESBL genes; either 2 or all the 3 genes occurred together. The Proteus isolates showed the presence of the TEM genes only [Table/Fig-5].
Distribution of TEM, SHV and CTX-M ESBL types among 93 study isolates.
| ESBL type | E.coli | Klebsiella pneumoniae | Proteus spp | Citrobacter | Total |
|---|
| TEM | 2 | | 8 | | |
| SHV | | 3 | | | |
| CTX-M | 35 | 6 | | | |
| CTX-M +SHV | 13 | 6 | | | |
| CTX-M + TEM | 5 | | | | |
| CTX-M + TEM + SHV | | 6 | | | |
| Nil | 4 | 3 | | 2 | |
| TOTAL | 59 | 24 | 8 | 2 | 93 |
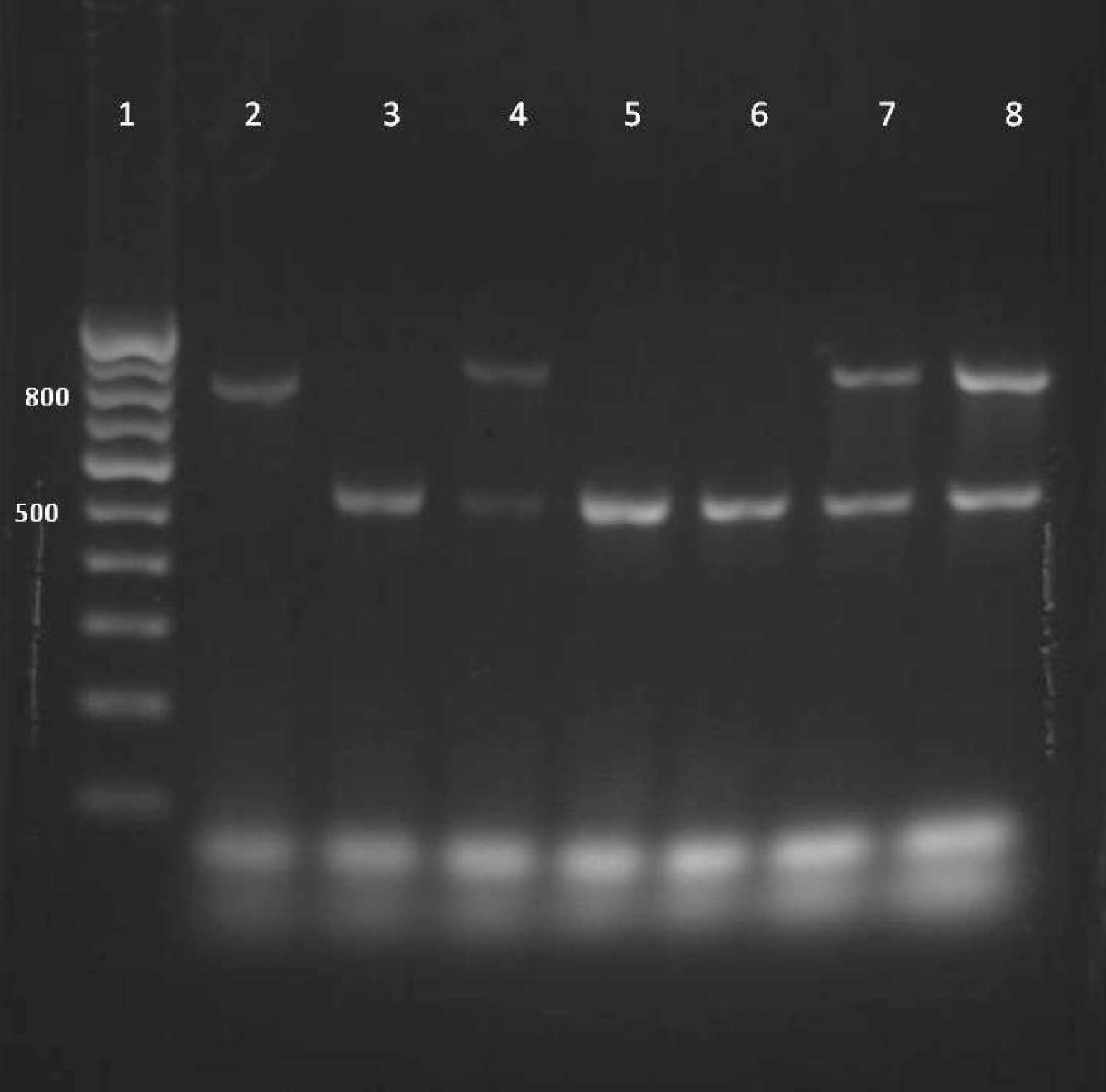
Agarose gel electrophoresis of PCR amplified products showing the presence of CTX-M (550bp), SHV (842bp) and TEM (918bp) genes in test and control strains.
Lane 1-DNA ladder
Lane 2- SHV positive control strain (Klebsiella pneumoniae ATCC 700603)
Lane 3 to lane 8-test strains
Lane 3,5,6- CTX-M
Lane 4,7,8- CTX-M + TEM
DISCUSSION
The emergence of plasmid mediated ESBLs among the members of Enterobacteriaceae have increased worldwide. The incidence of ESBLs in the major hospitals of India has been reported to be as high as 6-87% [7–9]. We reported 45.8% (229) ESBL producers by the CLSI confirmatory test. The maximum ESBL production was seen among the isolates of Klebsiella pneumoniae (52.27%), followed by E.coli (46.43%). Among the phenotypic methods which were used to find the ESBL production, the CLSI confirmatory method was found to be equally sensitive and cost effective as compared to the E-test, which had limited use in resource limited settings.
The present study also documents that the ESBL producers are also resistant to other non beta lactam antibiotics like fluroquinolones and aminoglycosides and this fact was statistically significant.
It was interesting to note that specific ESBLs appeared to be unique to a certain country or region. Though the prevalence of ESBLs has been recognized in various parts of the country, there is only limited data on its genotypes in this part of northern India. Our finding emphasized the increasing roles of the multiple ESBL genes in antibiotic resistance and this led to the consideration of an empirical treatment for the infections which were caused by coliforms, especially in patients who were compromised by an underlying disease or who had a compromised immunological status. The data which has been presented here, demonstrates the presence of the CTX-M genes in this part of our country. A similar study from south India also reported the CTX-M genes as the predominant ESBL genes [10]. CTX-M was reported to be the most prevalent gene among the E.coli strains and the Klebsiella species in another similiar study, but their co-production was not reported [11]. Coproduction of all the three genes, which was also seen in our study, was also reported in Klebsiella species in a recent study from south India [12].
It can be concluded from this study, that microorganisms have been evolving towards an antimicrobial drug resistance and this has been considered as a major challenge in hospitals. The use of broad spectrum antibiotics such as quinolones , the second, third and fourth generation cephalosporins, beta lactams combined with beta lactamase inhibitors and hospitalization are some of the risk factors for the infections which are caused by ESBL producing gram negative bacteria. There is a vicious circle between the antimicrobial resistance and the antimicrobial use. An indiscriminate use of the higher antibiotics should be restricted as far as possible. The infection control programmes should be monitored continuously in hospitals, to contain these ESBL producers.