Dengue is an endemic arboviral infection which affects the tropical and the subtropical regions around the world, predominantly the urban and the semiurban areas [1].
Dengue infects an estimated 50 to 100 million individuals and it is responsible for as many as 5,00,000 hospitalizations [2].
Since Dengue presents either as an asymptomatic or a mild undifferentiated fever in more than half of the infected individuals, the diagnosis has to be based on the clinical features, which has to be supported by positive serology and molecular tests.
There are 4 antigenically distinct serotypes of the Dengue viruses (DENV-1-4) that can cause human infections [3].
We are reporting the clinical, serological and the molecular investigations of a major Dengue like illness which was recorded at a tertiary care institute, between August to December 2007. Our study demonstrated a clear shift in the dominant serotypes from DENV-3 to DENV- 4 in this major outbreak.
MATERIALS AND METHODS
Clinical Samples
This was a prospective study which was done during the outbreak of Dengue in 2007.This study was approved by the institutional ethical committee. Seven hundred and thirteen (713) clinically suspected, consecutive cases of Dengue/DHF/DSS, who either reported directly or were referred to a tertiary care institute for treatment from the regions in and around Hyderabad, between August and December 2007, were included in the study. There was no sampling bias or any attempt to specially recruit patients for the study.
Of the 713 suspected cases, 590 (82.7%) were managed conservatively on an outpatient basis, while the remaining 123 (17.3%) cases needed hospitalization for management of their acute illness which was associated with the complications.
The acute phase serum samples were collected from all the 713 individuals by using serum separation BD vacutainers without EDTA (for the serology tests). Additional whole blood samples were collected from the 123 hospitalized patients by using BD vacutainers (Becton and Dickenson) with EDTA (for the molecular tests and the prospective virus isolation).
The Dengue/DHF/DSS case proformas which were prepared as per the WHO protocol, were filled in by the treating clinicians [9]. The case records of the 123 hospitalized cases were analyzed for the clinical and the laboratory data.
SEROLOGICAL ASSAYS
All the 713 sera were separated and tested on the same day for the anti IgG and the anti IgM Dengue antibodies by Dengue IgG capture ELISA and Dengue IgM capture ELISA (Pan Bio pvt. Ltd, Queensland, Australia).
MOLECULAR ASSAYS
The sera of 387 patients (264 outpatients and 123 hospitalized patients) and the plasma of 123 hospitalized patients, which had a positive anti Dengue serology, were further processed by molecular assays for the detection of the Dengue virus specific RNA.
Duplex RT-PCR
The viral RNA was extracted from 140μl of the serum/ plasma samples by using the QIAamp viral RNA mini kit (Qiagen, Germany). The presence of the Dengue/ Chikungunya specific RNA in the clinical samples was detected by using a Duplex RT-PCR (D-RT-PCR) [10]. Briefly, the PCR was carried out in a 25-μL reaction volume by using the Access quick one step RT-PCR kit (Promega, USA) which contained 2X PCR master mix, 2.5 U of the Avian Myeloblastoma Virus-Reverse Transcriptase (AMV-RT), and 20 pmol each of the DENV- and the CHIKV-specific primer pairs (DEN1, DEN2, CHIK1, and CHIK2). After their amplification, the PCR products were electrophoresed and visualized on a Bio Rad Gel Documentation system.
Nested PCR
Twenty Dengue RT-PCR positive amplicons were subjected to nested PCR through a second round of PCR for the Dengue virus serotyping [8]. Briefly the Oligonucleotide consensus primers were designed to anneal to any of the four Dengue virus types and to amplify a 511-bp product in a reverse transcriptase- Polymerase Chain Reaction (PCR). First, we produced a cDNA copy of a portion of the viral genome in a reverse transcriptase reaction, in the presence of primer D2 and then, we carried out a standard PCR (35 cycles of heat denaturation, annealing, and primer extension), with the addition of primer D1. The resulting double-stranded DNA product of the RT-PCR was typed by a second round of PCR amplification (nested PCR) with type-specific primers, which yielded DNA products, the unique sizes of which were diagnostic for each Dengue virus serotype.
The Dengue Virus Isolation
The virus isolation was attempted in C6/36 cell lines from all the 20 PCR positive plasma samples by following the standard virus adsorption technique [11]. The isolated viruses were confirmed by nested PCR and sequencing.
Nucleotide Sequencing and Phylogenetic Analysis
Nucleotide sequencing of the C-prM gene junction of 5 randomly selected Dengue viruses from the clinical samples, which included three DENV-4 (ND98, ND73, ND110) and two DENV-3 (ND14, ND143), was carried out by employing the Big Dye Terminator Cycle Sequencing Ready Reaction kit with an ABI 3100 sequencer (Applied Biosystems, USA) for identifying the genotype of the Dengue virus serotype by following the standard protocol [12]. The sequences were initially subjected to BLAST to find the closest sequence identity. Further, 2 phylogenetic analyses which were based on the C-prM gene junction of DENV-4 and DENV-3 were carried out by including a large number of geographically diverse DENV gene sequences, by using the Neighbour-Joining (NJ) method of the MEGA3 software version 3.1 [13].
The sequences of ND73 and ND110 were submitted to GenBank under the accession numbers, HM237348 and HM237349, respectively.
RESULTS
Of the 713 patients who were included in this 5 months study, the male (80) to female (43) ratio was 2:1. The most affected age group was the 21-30 years age group.
Among the 123 hospitalized patients whose case records were analyzed, the major clinical presentation was a febrile illness with or without skin rash and vomiting. The features of thrombocytopenia and elevated liver enzymes were also predominant in these patients, as has been shown in [Table/Fig-1].
Major clinical symptoms of hospitalized patients (n = 123) n is total number of cases.
| Clinical Symptoms | Number of Cases (%) |
|---|
| Fever | 100 |
| Thrombocytopenia | 77 |
| Maculopapular Rash | 31 |
| Elevated Liver Enzymes | 51 |
| Vomitings | 35 |
| Headache | 26 |
| Arthralgia/ Myalgia | 32 |
| Bleeding Manifestation | 25 |
| Pain Abdomen | 18 |
| Conjuctival Congestion | 10 |
| Respiratory Complaints | 11 |
| Neurological Complaints | 10 |
| Hypotension | 8 |
| Diarrhoea | 9 |
The serological analysis of the samples indicated 54% seropositivity (387/713 patients), with 18% IgM positivity (70 patients), 32% IgG positivity (123 patients) and 50% IgM and IgG positivity (194 patients). The serological tests were done by using IgM capture and IgG capture ELISA in which the cut off value of IgG was set to >1.79 to discriminate between the high levels of IgG (which were characteristic of the secondary Dengue infections) and the low levels of IgG (which were characteristic of the primary/past Dengue infections) [14]. By using this method, a majority of the secondary Dengue infections could be clearly delineated.
The secondary Dengue infections were more predominant in our study (79%). Five out of 387 (1.3%) progressed to the Dengue Shock Syndrome (DSS) and all these patients succumbed to the illness. The major clinical symptoms in the DSS cases were hypotension and shock. All the DSS cases had secondary Dengue infections.
Twenty out of the 387 (5.16%) serum samples were found to be positive for the Dengue-specific RNA through the demonstration of the 511-bp dengue complex-specific amplicon by D-RT–PCR. All these amplicons were further subjected to nested RT–PCR for their serotyping, which revealed 11 as DENV-4 positive and 9 as DENV-3 positive. The results of RT-PCR and ELISA have been shown in [Table/Fig-2].
Results of ELISA and PCR-positive samples
| Serotype | IgM+/IgG+ | IgM-/IgG+ | IgM+/IgG- | IgM-/IgG- | Total |
|---|
| Denv-3 | 2 | 5 | - | 2 | 9 |
| Denv-4 | 5 | 3 | - | 3 | 11 |
Eight out of the 387(2.06%) cases had dual infections with the Dengue and the chikungunya viruses, as was evidenced through the presence of both the DENV and the CHIKV RNA.
The case records of the 5/12 cases of DENV-4 who were hospitalized, were analyzed. The secondary infections, as per the serology, were predominant in these DENV-4 cases. None of the patients with DENV-3 needed hospitalization. The primary infections were predominant in the cases with the DENV-3 infections.
The major clinical findings in the cases with the DENV-4 infections have been shown in [Table/Fig-3].
Major clinical findings in cases with denv-4 infections. D.F = dengue fever.
| Clinical symptoms | No. of cases (%) | Clinical diagnosis |
|---|
| Febrile illness | 5 (100 %) | D.F |
| Thrombocytopenia | 4 (80 %) | D.F |
| Elevated liverenzymes | 4 (80 %) | D.F |
| Hepatitis | 2 (40 %) | D.F |
| Pain abdomen | 2 (40 %) | D.F |
| Vomitings | 2 (40 %) | D.F |
| Rash | 1 (20 % ) | D.F |
The 3 serial passages of the 20 acute phase samples resulted in the isolation of two DENV-4 (ND73, ND110) and one DENV-3 (ND143) isolates. The isolations were also confirmed by nested PCR and sequencing.
The nucleotide sequences of the C-prM gene junction (454bp; excluding the primer sequence) of the 5 representative dengue viruses (three DENV-4 and two DENV-3) were deciphered directly from the clinical samples. The pair-wise nucleotide sequence comparison revealed that all the 3 DENV-4 isolates from this outbreak were closely related (96.1-98.9% sequence identity).
These isolates from this outbreak revealed a nucleotide sequence identity of 94.7-95.8% with the prototype Philippines dengue-4 isolate (H-241) of 1956. A dendrogram was constructed after a pair-wise comparison of the 361 nucleotide sequences of the C-prM gene junction (position 156-516, with respect to H-241 (AY947539)), which classified all the isolates into three different genotypic groups [12,13,15]. The dendrogram revealed that the Indian Dengue-4 isolates from this outbreak belonged to the genotype I [Table/Fig-4]. This genotype was also represented by the isolates from a large number of countries, which included Thailand, Philippines, China, Sri Lanka and Brazil.
Phylogenetic tree among DENV-4, generated by Neighbor-Joining method based on the nucleotide sequence of C-prM gene junction. Each strain is abbreviated with the country of origin and last two digits of the year of isolation, followed by GenBank accession number in parenthesis. The DENV-4 sequenced in this study are indicated with a bold circle. Bootstrap values are indicated at the major branch points.
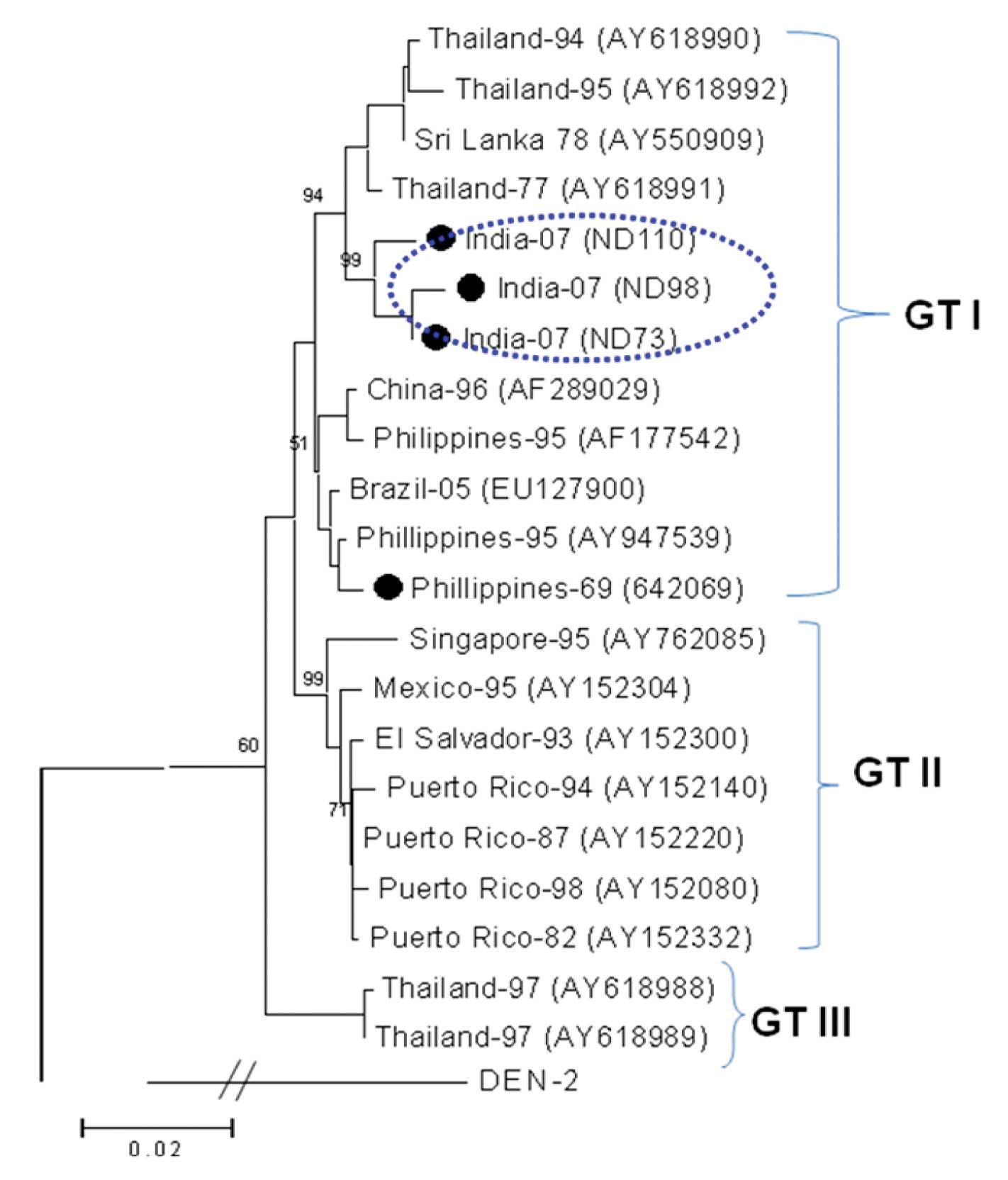
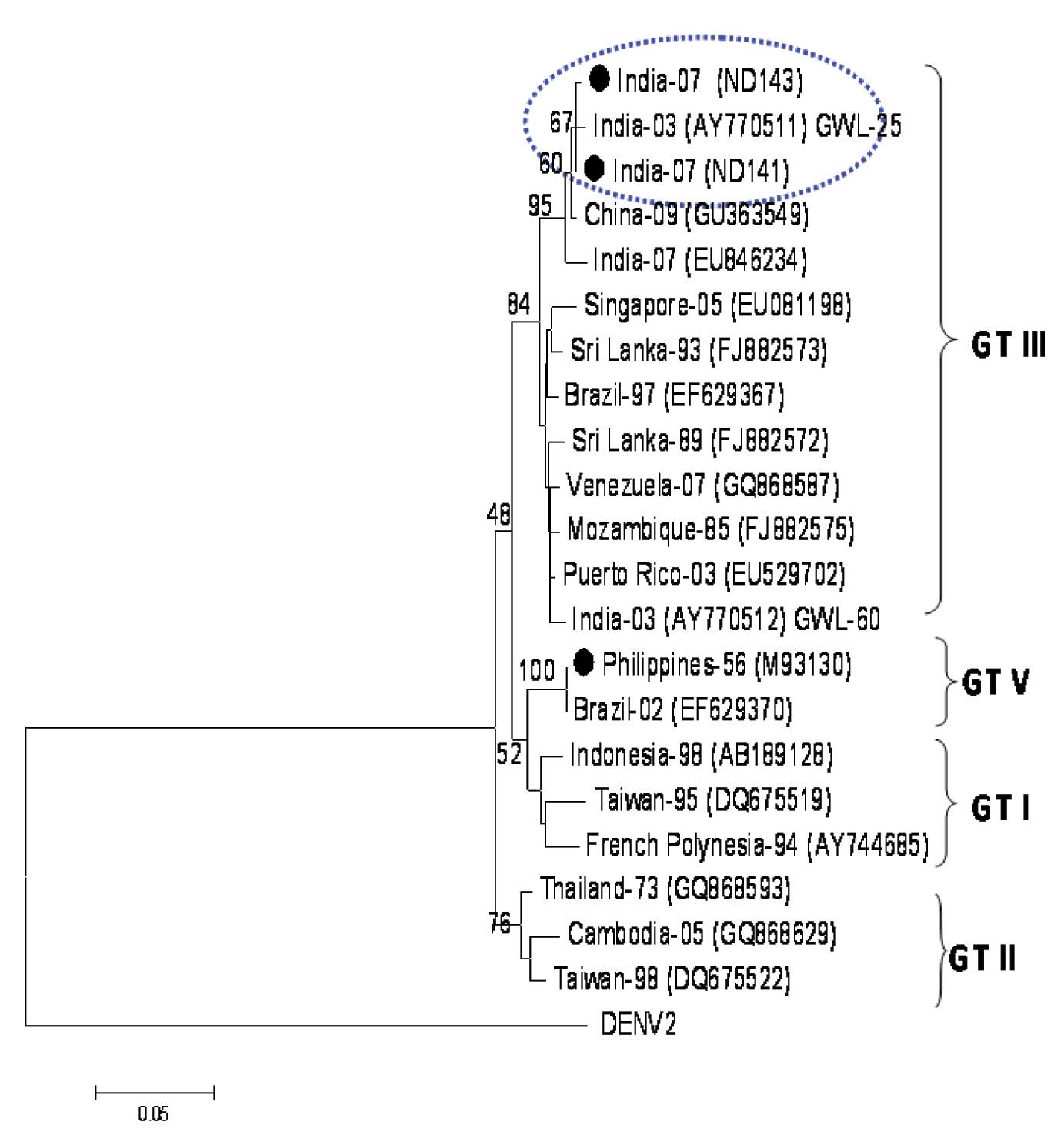
The two DENV-3 viruses which were sequenced in this study were found to be very closely related (99.8%). The BLAST search revealed a close identity (99%) with the earlier Dengue-3 viruses which had circulated in India since 2003. They also revealed a 94.5% sequence identity with the prototype DEN-3 isolate (H-87). The phylogenetic analysis grouped these DENV-3 viruses into genotype III, along with the circulating Indian DENV-3 viruses [Table/Fig-5]. This genotype is cosmopolitan in nature and it is represented by the isolates from Asia, Africa and the American continents.
Phylogenetic tree among DENV-3, generated by Neighbor-Joining method based on the nucleotide sequence of C-prM gene junction. Each strain is abbreviated with the country of origin and last two digits of the year of isolation, followed by GenBank accession number in parenthesis. The DENV-3 sequenced in this study are indicated with a bold circle. Bootstrap values are indicated at the major branch points.
DISCUSSION
Though Dengue infections were reported in India since the late 1950s, an upsurge in its activity has been noticed since the mid 1990s.
The first isolation of the Dengue virus serotypes 1and 4 was reported from India in 1964, and that of the Dengue virus serotype 3 was reported in 1996 [16, 17].
Our data showed that the epidemic waves were seen in the urban centres, because the highly urbanized areas had substantial proportions of the populations who lived in crowded and impoverished areas with poor sanitation [18].
The transmission of Dengue increases in the monsoon and the post monsoon seasons, as was also observed in our study. Young adults were found to be more affected and a male preponderance was observed in our study.
The most common clinical symptoms which were seen among the study cases were fever, thrombocytopenia, rash and elevated liver enzymes. The association of the Dengue parameter positivity with thrombocytopenia was in concordance with the findings of another recent Indian study which was done by R.D.Kulkarni et al., [19]. Dengue hepatitis was observed in 2 patients with the DENV-4 infection, which was not observed in our previous study [18].
All the 5 cases that progressed to DSS, expired due to secondary infections. In the current study, the demonstration of the Dengue RNA in 5.16% of the samples by RT–PCR and the detection of the IgM antibodies in 18% of the samples, confirmed the causative agent of this outbreak as the Dengue virus.
The isolation of the DENV-4 and the DENV-3 viruses from the clinical samples further confirmed this aetiology. The close branching pattern of the DENV-4 viruses with the Asian viruses from Sri Lanka and Thailand indicated the circulation of similar viruses in the Asian neighborhood. Though the association of DENV-4 in the major Indian Dengue outbreaks has rarely been reported, however, its silent circulation may not be ruled out, keeping in mind its circulation in the neighbouring countries. The identification of the closely related DENV-3 (genotype III) in this outbreak indicated its continued circulation in south India. 42% of the cases with the DENV-4 infection needed hospitalization and all of them were managed conservatively without an intensive care treatment. This was in contrast to the findings of the study which was done by Cecilia et al., [20] where both the DENV-4 cases needed intensive care treatment and one case had expired. All the cases with DENV-4 in our study were clinically diagnosed with Dengue fever and they were managed conservatively; no mortality was recorded among them. 25% of the cases with DENV-3 needed hospitalization. The major clinical symptoms among them were fever, elevated liver enzymes, diarrhea and vomiting and all of them were managed conservatively.
We recorded a co-infection of Chikungunya along with Dengue in 2.06% of the cases. This was in concordance to the findings of the study which was done by Chahar HS et al., [21]. All the patients with this co-infection had fever, headache, joint pain and low thrombocyte counts (<100,000/mm3) and all of them were managed conservatively.
Since the clinical features of DENV and CHIKV are usually similar, the CHIKV infections may go undiagnosed in the DENV-endemic areas. In India, the Aedes aegypti mosquitoes are the primary vectors for DENV and CHIKV, and the opportunities for co-infections in humans are increasing, due to the feeding behaviour of the mosquitos [21], the low socioeconomic conditions, and the high population density. Serologic investigations which were done in southern India indicated that the two viruses could co-exist in the same host [22]. In the Indian setting, while screening, considering both the Dengue and the chikungunya infections is necessary, because though the clinical features are similar, the outcomes may be different [23].
This study clearly indicated the sudden dominance of DENV-4 in an Indian Dengue outbreak. The identification of a dual infection with CHIKV also indicated the co-circulation of the Dengue and the CHIKV viruses in an area. A prompt detection of the aetiology, coupled with an effective patient management, can play a very important role in the outcome of the Dengue infection.
CONCLUSION
The genetic characterization of the Indian DENV-4 and DENV-3 and its correlation with the clinical outcome provided important information for the future molecular diagnostic investigations. The surveillance of the Dengue viruses needs to be closely monitored for the emergence of newer serotypes in hitherto unknown areas.