Use of Triplex PCR for Rapid Detection of PVL and Differentiation of MRSA from Methicillin Resistant Coagulase Negative Staphylococci
Nagarajan Abimanyu1, Arunkumar Krishnan2, Saravanan Murugesan3, Kaushik Subramanian G4, Sivakumar Gurumurthy5, Padma Krishnan6
1 Department of Microbiology, Dr. ALM PG Institute of Basic Medical Sciences, University of Madras, Taramani, Chennai, India – 113.
2Madras Medical College and Govt. General Hospital, Park Town, Chennai, India – 2.
3 Dept. of Microbiology, Dr. ALM PG Institute of Basic Medical Sciences, University of Madras, Taramani, Chennai, India – 113.
4 Dept. of Microbiology, Dr. ALM PG Institute of Basic Medical Sciences, University of Madras, Taramani, Chennai, India – 113.
5Madras Medical College and Govt. General Hospital, Park Town, Chennai, India – 2.
6 Dept. of Microbiology, Dr. ALM PG Institute of Basic Medical Sciences, University of Madras, Taramani, Chennai, India – 113.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Padma Krishnan, Assistant Professor, Dept of Microbiology, Dr. ALM PG Institute of Basic Medical Sciences, University of Madras, Taramani, Chennai, India – 113.
Phone: +919840742105, 044 – 24547105 Fax: 044-24540709
E-mail: padma.abpkn@gmail.com
Introduction: Methicillin-Resistant Staphylococcus aureus (MRSA) has become a major public health problem in both hospitals and communities. Panton – Valentine Leucocidin (PVL) has been reported to be an important marker for the highly pathogenic community acquired S. aureus infections. A rapid detection of these MRSA is very important for its treatment. The specific detection of MRSA is always a problem due to the prevalence of methicillin resistance among the coagulase negative Staphylococci. Hence, this study was done to develop a rapid triplex PCR for the detection of PVL positive MRSA and for the simultaneous differentiation of MRSA from Coagulase Negative Staphylococci (CoNS).
Materials and Methods: We developed a triplex PCR for the specific detection of PVL positive Community Acquired (CA) – MRSA and for its simultaneous differentiation from the coagulase negative Staphylococci. We used PCR for targeting the fem A gene which is specific for S. aureus, mecA which is specific for methicillin-resistance and luk - PV which is specific for the PVL toxin. The method was evaluated with a total of 100 clinical isolates of Staphylococcus spp.
Results: The triplex PCR was successfully standardized by using the reference strains and it was evaluated by using clinical strains. The method was found to be rapid, highly sensitive (100%), specific (99%) and cost effective.
Conclusion: Triplex PCR can be used as a diagnostic tool for the detection of the highly pathogenic strains of CA-MRSA.
PVL MRSA, MRCoNS, Triplex PCR, femA, mecA
INTRODUCTION
Staphylococcus aureus is a major human pathogen which causes various infections, both in the otherwise healthy individuals and in hospitalized patients. Over 60% of the clinical isolates of S. aureus were found to produce the penicillin binding protein 2a(PBP2a)- the factor which is responsible for the methicillin resistance, which is encoded by the gene, mecA [1]. The mecA gene has also been reported in coagulase negative staphylococcus (CoNS). About 62% of the CoNS, (S. epidermidis (75%), S. lugdunensis (25%), S.hemolyticus (12%) and S. saprophyticus (5%) have been reported to be methicillin-resistant [2]. femA is a chromosomally encoded ubiquitous gene in Staphylococci, which is involved in the formation of the peptidoglycan pentaglycine interpeptide linkages [3]. It also plays a role in the expression of methicillin resistance, the mechanism of which is not clearly understood. femA can be used as a marker for the differentiation of S. aureus from the coagulase negative Staphylococci. Panton- Valentine leukocidin (PVL) has gained much importance in the recent past due to its association with the community acquired methicillin-resistant S. aureus (CA-MRSA) infection. is a phage encoded exotoxin of S. aureus and it has been found to be cytotoxic to rabbit and human neutrophils as it induces apoptosis. The S. aureus isolates with PVL are rapidly spreading and they cause serious skin and soft tissue infections such as pyomyositis, abscesses, breast abscesses, necrotizing fasciitis and pneumonia in otherwise healthy individuals [4–8]. Molecular methods are available for the detection of methicillin-resistance among the Staphylococcus spp. [9–11]. But, only few methods are available for the detection of PVL and for the simultaneous identification and differentiation of MRSA and methicillin-resistant coagulase negative Staphylococci (MRCoNS) [12, 13]. Hence, in this study, a rapid triplex PCR which targetted the femA, mecA and the pvl genes was used for the specific detection of PVL and methicillinresistance and for the differentiation of MRSA from MRCoNS.
MATERIALS AND METHODS
The bacterial isolates
The method was standardized by using the 10 standard strains of Staphylococci, which included 6 MRSA, 2 S. aureus and 2 CoNS [Table/Fig-1].
List of reference strains used for the standardization of the Triplex PCR.
| S. aureus ATCC 29213 | MSSA | femA |
| S. aureus ATCC 25923 | MSSA | femA, pvl |
| S. aureus ATCC 43300 | MRSA | femA, mecA |
| S. aureus COL | MRSA | femA, mecA |
| MRSA-Mu50 | MRSA | femA, mecA |
| MRSA-Mu3 | MRSA | femA, mecA |
| MRSA-USA300 (FPR3757) | MRSA | femA, mecA, pvl |
| MRSA-USA400 (MW2) | MRSA | femA, mecA, pvl |
| S. epidermidis ATCC 12228 | MRSE | mecA |
| S. epidermidis ATCC 35984 | MRSE | mecA |
A total of 100 retrospective clinical isolates of Staphylococci, which were previously confirmed by standard phenotypic methods, were used for the evaluation of the triplex PCR. The bacterial isolates included 25 clinical isolates of methicillin-susceptible S. aureus (MSSA), 25 clinical isolates of MRSA, 25 clinical isolates of CoNS and 25 clinical isolates of MRCoNS. The clinical isolates were collected between June to August 2011 from a tertiary care centre in Chennai, from various clinical specimens. An institutional ethical clearance was obtained and an informed consent was obtained from the study participants. The isolates were identified by standard methods [14]. The methicillin-resistance was detected by cefoxitin disc diffusion and oxacillin agar dilution methods [14].
The triplex PCR method
The triplex PCR method for the detection of PVL and the simultaneous differentiation of MRSA from MRCoNS was designed by using the primer sequences from various published studies [Table/Fig-2] [4, 15, 16]. The concentrations of the forward and reverse primers of each gene (pvl – 0.1μM each, mecA – 0.2μM each and femA – 0.08μM each) were optimized for the multiplex PCR.
Multiplex PCR method for detection of pvl , MRSA and differentiation from MRCoNS.
| Primer sequences | PCR Reaction Mixture (25μl reaction volume) | PCR cycling conditions (Eppendorf Gradient Mastercycler) | Amplicon separation by 1.5% agarose gel electrophoresis |
|---|
| femA (Mehrotraet al., 2000) [23] F: 5′ – AAAAAAGCAC ATAACAAGCG – 3′ R: 5′ – GATAAAGAAGA AACCAGCAG – 3′ | 10X Taq polymerase buffer; 2.5U Taq DNA polymerase (NEB); 200mM dNTPs (Bio-tools); 10pico moles of each primer (Sigma) | Intial Denaturation: 94oC/ 2min | 132bp |
| mecA (Kondo et al., 2007) [24] F: 5′-TGCTATCCACC CTCAAACAGG-3′ R: 5′-AACGTTGTAAC CACCCCAAGA-3′ | 30 cycles of Denaturation: 94oC/ 45sec; Annealing: 55oC/30sec; Extension: 72oC/45sec | 286bp |
| pvl (Lina et al.,1999)[6] F:5′–ATCATTAGGTAAAAT GTCTGGACATGATCCA–3′ R: 5′– GCATCAASTGTATT GGATAGCAAAAGC– 3′ | Final Extension: 72oC/ 2min | 441bp |
The template DNA for the multiplex PCR was prepared from an overnight culture (Brain Heart Infusion Agar), which was obtained by boiling a few colonies of Staphylococci in 100μl of DNase free water (Qiagen, Germany) for 10 minutes and centrifuging the suspension at 10,000 rpm for 3min. Five micro-litres of the supernatant was used as the template for the PCR. The PCR was carried out in a 25μl reaction mix which contained 200μM of dNTPs (NEB), 1X PCR buffer (NEB) and 0.5U Taq DNA polymerase (NEB). The amplification was done by using a Mastercycler Gradient (Eppendorf, Hamburg, Germany) under the following cycling conditions (one cycle of initial denaturation at 94°C for 4 min, 25 cycles of denaturation at 94°C for 30s, annealing for 30s at 54°C, and extension at 72°C for 30 s, followed by a final extension at 72°C for 5 min. The amplified products were separated on a 1.5% low melting agarose gel (Medox India Pvt Ltd). They were electrophoresed (0.5X TBE buffer at 150V and 90mA for 30 minutes), stained with 0.5% ethidium bromide, visualized and recorded by using gel documentation system (BIO-RAD). A 100bp ladder (RBC Bioscience Corp, Taiwan) was run as a molecular marker.
Standardization of the triplex PCR for the detection of PVL MRSA
In this study, the triplex PCR was optimized to specifically identify the S. aureus at the species level (femA) and to detect the methicillin- resistance (mecA) and the PVL toxin (pvl) simultaneously by using the previously described primers in three different studies. Standardization of the PCR was done by a stepwise optimization of the individual reaction components: the primers, MgCl2 and the dNTPs. The annealing temperature for the triplex PCR was standardized by a gradient technology by using the Master Cycler Gradient (Eppendorf, Hamburg, Germany). The Triplex PCR was found to produce excellent results with 1.5mM MgCl2, 0.5U Taq DNA polymerase, 200μM dNTPs, 10ng of DNA and a 55°C annealing temperature. The specificity of the triplex PCR was determined by using 10 reference strains of Staphylococci, which included 6 MRSA, 2 MSSA and 2 methicillin-resistant S. epidermidis (MRSE). To test the specificity of the triple PCR assay, the DNA templates from Escherichia coli ATCC25924 and Enterococcus faecalis ATCC 29212 were included. As a negative control, the reaction mixture was tested with sterile water.
RESULTS
FemA was amplified in all the standard strains of S. aureus, which included 6 MRSA and 2 MSSA, whereas no amplification was seen in S. epidermidis. All the methicillin-resistant Staphylococci, which included 6 MRSA and 2 MRSE, were found to be positive for the mecA gene, whereas both the MSSA isolates were negative for the mecA gene. The reference strains of S. aureus, ATCC25923, MRSA USA300 FPR3757 and MRSA MW2 were positive for pvl, whereas all the other reference strains were found to be negative. The representative picture of the agarose gel electrophoresis has been shown in [Table/Fig-3].
PCR for detection of PVL and differentiation of MRSA from MRCoNS.
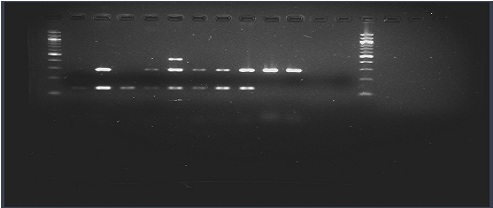
Evaluation of the Triplex PCR for the detection of PVL MRSA
After the standardization with the reference strains, the triplex PCR method was evaluated with 100 clinical strains of Staphylococci, which included 25 isolates of MRSA, 25 isolates of MSSA, 25 isolates of MRCoNS and 25 isolates of CoNS with a known methicillin resistance status. All the 25 isolates of MRSA were found to be positive for both femA and mecA. Also, 10/25 (40%) MRSA isolates were found to harbour the pvl gene, which is the marker for the highly pathogenic community acquired MRSA (USA300, MW2). of the 25 MSSA isolates, femA was detected in all the isolates and 3/25 (12%) isolates were found to be positive for pvl. One S. aureus isolate which was identified as MSSA by the phenotypic method was found to be positive for the mecA gene. The mecA gene alone was detected in all the 25 MRCoNS isolates which were included in the triplex PCR, whereas none of the genes was amplified for the methicillin-sensitive CoNS [Table/Fig-4]. There was a good correlation between the results of the triplex PCR assay and those of the traditional phenotypic methods.
Genes detected among the isolates used for evaluation of the Triplex PCR.
| Various genes detected | Number (%) of isolates |
|---|
| MRSA | MSSA | MRCoNS | CoNS |
| femA | 25 (100) | 25 (100) | 0 (0) | 0 (0) |
| mecA | 25 (100) | 1 (4) | 25 (100) | 0 (0) |
| pvl | 10 (40) | 3 (12) | 0 (0) | 0 (0) |
DISCUSSION
The genus, Staphylococcus comprises about 34 different species and methicillin-resistance was reported in most of the species, which included the most pathogenic species-S. aureus and other commensal species. The use of rapid molecular methods, which included PCR for the specific identification of S. aureus and the detection of methicillin-resistance, had been described previously [9, 10, 12, 13]. Most of the previous studies had used anyone of the following genes viz., nuc, 16S rDNA, coag and femA with mecA for the rapid and the specific detection of MRSA at the species level. The emergence of highly pathogenic CA-MRSA infections in healthy individuals [17] and their recent emergence as nosocomial pathogens [18, 19] with multidrug resistance, had necessitated the need of a rapid diagnostic technique for the specific detection of MRSA. The presence of pvl was considered as the marker for the CA-MRSA infection in most of the cases. Even though there are reports on CA-MRSA without pvl, its association with CA-MRSA has been reported to cause life threatening infections which include necrotizing pneumonia, necrotizing fasciitis, septicaemia, pyomyositis, brain abscess and pyogenic abscesses [4–8]. PVL is a necrotizing cytotoxin which is specific for the human and rabbit polymorphonuclear cells [20], which may be responsible for the Staphylococcal invasiveness and virulence. In developing countries like India [19, 21], Africa [22] and other Asian countries [23], the prevalence rate of the pvl positive CA-MRSA is very high as compared to those at other geographic locations [17, 24]. Even in this study, about 12% of the MSSA and 40% of the MRSA were found to be positive for PVL, which was slightly higher than that which was reported from other geographical regions and lower than the reports from Africa and India. Although pvl was extensively studied and considered as a major factor in the CA-MRSA infection, only few molecular methods employed pvl for the detection of the pathogenic CA-MRSA isolates [12,13]. We carefully selected the published primers from various studies and designed a triplex PCR which targetted pvl, mecA and femA. The above PCR method was standardized by using the reference strains and it was evaluated with 100 clinical isolates of Staphylococcus spp. Negative controls without the template were run and the DNA from other genera were included to rule out the false positive results which were due to the non specific amplification. The PCR successfully amplified all the 3 target genes and it was found to be highly specific (99%) and sensitive (100%). Only one S. aureus isolate which was identified as MSSA by the phenotypic method was found to be positive for mecA. This may be due to the negative expression of the gene in-vitro, as has been reported in earlier studies [25]. The current method which targetted femA, mecA and pvl can be used in resource-limited settings as a simple, rapid and a highly sensitive molecular tool for the detection of the life threatening CA-MRSA infections.
CONCLUSION
We successfully developed a new triplex PCR which can be used as a simple, rapid and a highly specific molecular diagnostic tool for the detection of the highly pathogenic strains of CA-MRSA, especially in developing countries like India.
[1]. Hiramatsu K, Cui L, Kuroda M, Ito T, The emergence and evolution of methicillin-resistant Staphylococcus aureusTrends Microbiol 2001 9:486-93. [Google Scholar]
[2]. Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997 – 1999Clin Infec Dis 2001 32(suppl 2):S114-S132. [Google Scholar]
[3]. Berger-Bachi B, Barberis-Maino L, Strassle A, Kayser FH, femA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterizationMol Gen Genet 1989 219:263-69. [Google Scholar]
[4]. Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumoniaClin Infect Dis 1999 29:1128-32. [Google Scholar]
[5]. Boyle-Vavra S, Daum RS, Community-acquired methicillin-resistant Staphylococcus aureus: the role of Panton-Valentine LeukocidinLab Invest 2007 87(1):3-9. [Google Scholar]
[6]. Girish BK, Pal PK, Veenakumari HB, Goyal M, Kovoor JME, Savitha N, Community acquired methicillin-resistant Staphylococcus aureus pyomyositis with myelitis: A rare occurrence with diverse presentationNeurology India 2009 57(5):653-56. [Google Scholar]
[7]. Gayathri S, Indira J, Boil to sepsis: Case of community acquired MRSAIndian Pediatrics 2009 46(6):537-38. [Google Scholar]
[8]. Nagarajan A, Arunkumar K, Saravanan M, Sivakumar G, Krishnan Padma, PVL positive methicillin-resistant Staphylococcus aureus breast abscess infection among post-partum women in Chennai, South IndiaBMC Infect Dis 2012 12(Suppl 1):O13 [Google Scholar]
[9]. Murakami K, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S, Identification of methicillin-resistant strains of staphylococci by polymerase chain reactionJ Clin Microbiol 1991 29(10):2240-44. [Google Scholar]
[10]. Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G, Specific detection of methicillin-resistant Staphylococcus species by multiplex PCRJ Clin Microbiol 1995 33(11):2864-67. [Google Scholar]
[11]. Maes N, Magdalena J, Rottiers S, De Gheldre Y, Struelens MJ, Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative Staphylococci and determine methicillin resistance from blood culturesJ Clin Microbiol 2002 40(4):1514-17. [Google Scholar]
[12]. McClure JA, Conly JM, Lau V, Elsayed S, Louie T, Hutchins W, Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from -resistant staphylococciJ Clin Microbiol 2006 44(3):1141-44. [Google Scholar]
[13]. Al-Talib H, Chan YY, Alyaa A, Khateeb HH, Kirnpal-Kaur BS, Karim AJ, A pentaplex PCR assay for the rapid detection of methicillinresistant Staphylococcus aureus and Panton-Valentine LeucocidinBMC Microbiol 2009 9:113 [Google Scholar]
[14]. Crossely KB, Jefferson KK, Archer GL, Fowler Jr VG, Staphylococci in human disease 2009 SecondWiley Blackwell publishing:235-52. [Google Scholar]
[15]. Mehrotra M, Wang G, Johnson WM, Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1 and methicillin resistanceJ Clin Microbiol 2000 38(3):1032-35. [Google Scholar]
[16]. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Combination of multiplex PCRs for Staphylococcal Cassette Chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regionsAntimicrob agents and chemother 2007 51(1):264-74. [Google Scholar]
[17]. Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergenceEmerg Infect Dis 2003 9:978-84. [Google Scholar]
[18]. Maree CL, Daum RS, Boyle-Vavra S, Matayoshi K, Miller LG, Community- associated methicillin-resistant Staphylococcus aureus isolates which cause healthcare-associated infectionsEmerg Infect Dis 2007 13(2):236-42. [Google Scholar]
[19]. Nagarajan A, Ananthi M, Krishnan P, Reischl U, Prabha C, Lindeb H-J, Emergence of Panton Valentine leucocidin among community and hospital associated methicillin-resistant Staphylococcus aureus in Chennai, south IndiaJ Hosp Infect 2010 76(3):269-71. [Google Scholar]
[20]. Löffler B, Hussain M, Grundmeier M, Brück M, Holzinger D, Georg V, Staphylococcus aureus Panton-Valentine leukocidin is a very potent cytotoxic factor for human neutrophilsPLoS Pathog 2010 6(1):e1000715 [Google Scholar]
[21]. Mandelia C, Shenoy S, Community associated methicillin-resistant Staphylococcus aureus in skin and soft tissue infectionsJ Clin Diagn Res 2010 4:2673-77. [Google Scholar]
[22]. Okon KO, Patrick B, Auwalu U, Johnson L, Bukola O, Adebayo OS, Co-occurrence of predominant Panton-Valentine leukocidin-positive Sequence Type (ST) 152 and multidrug-resistant ST 241 Staphylococcus aureus clones in Nigerian hospitalsJ Clin Microbiol 2009 47(9):3000-03. [Google Scholar]
[23]. Hsu LY, Koh TH, Kurup A, Low J, Chlebicki MP, Tan BH, High incidence of Panton-Valentine leukocidin-producing Staphylococcus aureus in a tertiary care public hospital in SingaporeClin Infect Dis 2005 40(3):486-89. [Google Scholar]
[24]. Wannet WJ, Spalburg E, Heck ME, Pluister GN, Tiemersma E, Willems RJ, Emergence of virulent methicillin-resistant Staphylococcus aureus strains carrying Panton-Valentine leucocidin genes in The NetherlandsJ Clin Microbiol 2005 43:3341-45. [Google Scholar]
[25]. Swenson JM, Tenover FC, Results of disk diffusion testing with cefoxitin correlate with presence of mecA in Staphylococcus sppJ Clin Microbiol 2005 43(8):3818-23. [Google Scholar]