The bacterial resistance to the MLSB group may be expressed through different mechanisms which include target site modification, the macrolide efflux pump and enzymatic antibiotic inactivation. The modification of the ribosomal target is encoded by a multiallele plasmid borne erythromycin ribosome methylase (erm) gene that causes the production of the methylase enzymes. These enzymes cause methylation of the A2058 residue which is located in the conserved domain V of the 23S rRNA component of the 50S ribosomal subunit, which leads to cross resistance and the formation of the phenotype of the resistance pattern, called as the MLSB – resistant phenotype [7,8].
The resistance to the MLSB antibiotics can be either constitutive (MLSBc) or inducible (MLSBi). If the erm genes are consistently expressed, the organisms may show in vitro resistance to erythromycin (ER), clindamycin (CL) and to other members of the MLSB group and they are said to be of the MLSBc phenotype. However, if the erm genes require an inducing agent to express the resistance to CL, then the organisms are said to be of the MLSBi phenotype. The organisms which belong to the MLSBi phenotype are resistant to ER and sensitive to CL in vitro. The CL therapy in such patients can lead to clinical failures [9–12]. Low levels of ER is an inducer of the MLSBi phenotype and this forms the basis of the D-test [13].
Staphylococci can also develop macrolide resistance, based on the presence of the efflux pump which is encoded by the macrolide streptogramin resistance (msrA) gene, leading to resistance to the macrolides and the type B streptogramins, but not to the lincosamides. These isolates are known to be of the MS phenotype and they show in vitro resistance to ER and susceptibility to CL. But the CL therapy can safely be given in infections which are caused by the organisms of this phenotype and there is no risk of clinical failures [1]. Therefore, it is important to differentiate these two mechanisms of resistance.
It is very important that the clinical microbiologists and the infectious disease experts keep a close watch on the developing patterns of drug resistance, which will help in guiding the therapy effectively [14]. The Clinical Laboratory Standards Institute (CLSI) [15] has recommended the ER-CL disc approximation test (D-zone test) to detect the inducible clindamycin resistance. No previous data are available from this part of the state. This study was therefore taken up to close this gap in our knowledge. The aim of the present study was to detect the prevalence of inducible clindamycin resistance among the clinical isolates of Staphylococci.
MATERIALS AND METHODS
This was a cross sectional study which was conducted in the Department of Microbiology and Immunology, Veer Chandra Singh Garhwali Govt. Medical Sciences and Research Institute, Srikot, Uttarakhand, from July 2010 to December 2011. A total of 373 consecutive, non duplicate strains of Staphylococci isolated from various clinical samples like pus, wound swab, blood, urine and other body fluids, were tested.
The isolates were identified by using conventional biochemical reactions (catalase, coagulase, DNAse, etc.,) and they were tested for antibiotic susceptibility by the Kirby Bauer disc diffusion method on Muller Hinton agar (Hi-media Laboratories Pvt. Ltd, India). A suspension of the isolated colonies of each test strain equivalent to a 0.5 McFarland’s standard was prepared in sterile normal saline. Using a sterile cotton swab, the standardized inoculum was inoculated on Muller Hinton agar (MHA) plates. All the antibiotic susceptibility tests were interpreted in accordance with the CLSI guidelines [15].
Methicillin resistance was detected by taking cefoxitin as a surrogate marker. The clindamycin (CL, 2μg), erythromycin (ER, 15μg), and the cefoxitin (CF, 30μg) discs were procured from Hi-media Laboratories Pvt. Ltd, India. The quality control for the ER, CL, and the CF discs was performed with S. aureus ATCC 25923 (American Type Culture Collection, Manassas, VA, USA).
134 isolates of Staphylococci were selected, based on the discordant resistance pattern for ER and CL (ER-resistant and CL-sensitive) and they were subjected to the D-zone test for inducible clindamycin resistance as per the CLSI guidelines [15]. The ER disc was placed 15mm apart, edge to edge from the CL disc on the inoculated MHA plate and was incubated at 37°C. After incubation the plates were examined by using transmitted light to detect any flattening or blunting of the shape of the CL zone. The organisms that showed flattening or blunting of the zone of inhibition (D-shaped) around CL in the area between the two discs where both the drugs had diffused after 18-24 hrs of incubation, indicated inducible clindamycin resistance and they were designated as D-test positive. The organisms which showed absence of the blunted zone of inhibition were designated as D-test negative.
Three different phenotypes were interpreted as [16]:
1. The constitutive MLSB phenotype (MLSBc): The Staphylococcal isolates resistant to both ER (zone of inhibition ≤13mm) and CL (≤14mm).
2. the Inducible MLSB phenotype (MLSBi): The Staphylococcal isolates which showed resistance to ER (≤13mm) and sensitivity to CL (≥21mm) with a D-shaped zone of inhibition around the CL disc.
3. The MS phenotype: The isolates which showed a circular zone of inhibition around CL (≥21mm) and resistance to ER (≤13mm).
The results were photographed for records.
RESULTS
Among the 373 staphylococcal isolates 239 (64.1%) were S.aureus and 134 (35.9%) were coagulase negative staphylococci (CONS). The sample sources and the categorization of the isolates have been depicted in [Table/Fig-1].
Sample source and their categorization
| Source of Sample | Total No. of Sample | MSSA | MRSA | MSCONS | MRCONS |
| Pus | 173 | 136 | 08 | 22 | 07 |
| Wound Swab | 35 | 19 | 07 | 07 | 02 |
| Blood | 75 | 26 | 08 | 28 | 13 |
| Urine | 55 | 11 | 03 | 31 | 10 |
| Sputum | 27 | 12 | 05 | 06 | 04 |
| Others* | 08 | 04 | 0 | 02 | 02 |
| Total | 373 | 208 | 31 | 96 | 38 |
MRSA = methicillin resistant S.aureus, MSSA = methicillin sensitive S.aureus, MRCONS = methicillin resistant coagulase negative staphylococci, MSCONS = methicillin sensitive coagulase negative staphylococci *Pleural Fluid, Knee Aspirate, Semen, Vaginal discharge
192 (51.5%) isolates were susceptible to both CL and ER (CL-S, ER-S) and they were of the sensitive phenotype [Table/Fig- 2(a)]. 47(12.6%) strains showed constitutive resistance and they belonged to the MLSBc phenotype [Table/Fig-2(b)].; they showed in vitro resistance to both the drugs (CL-R, ER-R). The remaining 134 (35.9%) isolates expressed CL-ER discordant (CL-S, ER-R) results. Among the discordant isolates, 45 (33.6%) had the MLSBi phenotype [Table/Fig-2(c)].; and 89 (66.4%) had the MS phenotype [Table/Fig-2(d)]. The phenotypic characterization of the isolates has been depicted in [Table/Fig-3]. Among these, 45 isolates 6 (13.3%) were MRSA, 13 (28.9%) were MSSA and 26 (57.8%) were CONS. Overall, 45 (12.1%) out of the 373 staphylococcal isolates which were included in this study were reported as being resistant to CL, based on the D-test findings. Such a resistance would otherwise have been missed out on the routine Kirby-Bauer disc diffusion method.
Photograph showing different phenotypes
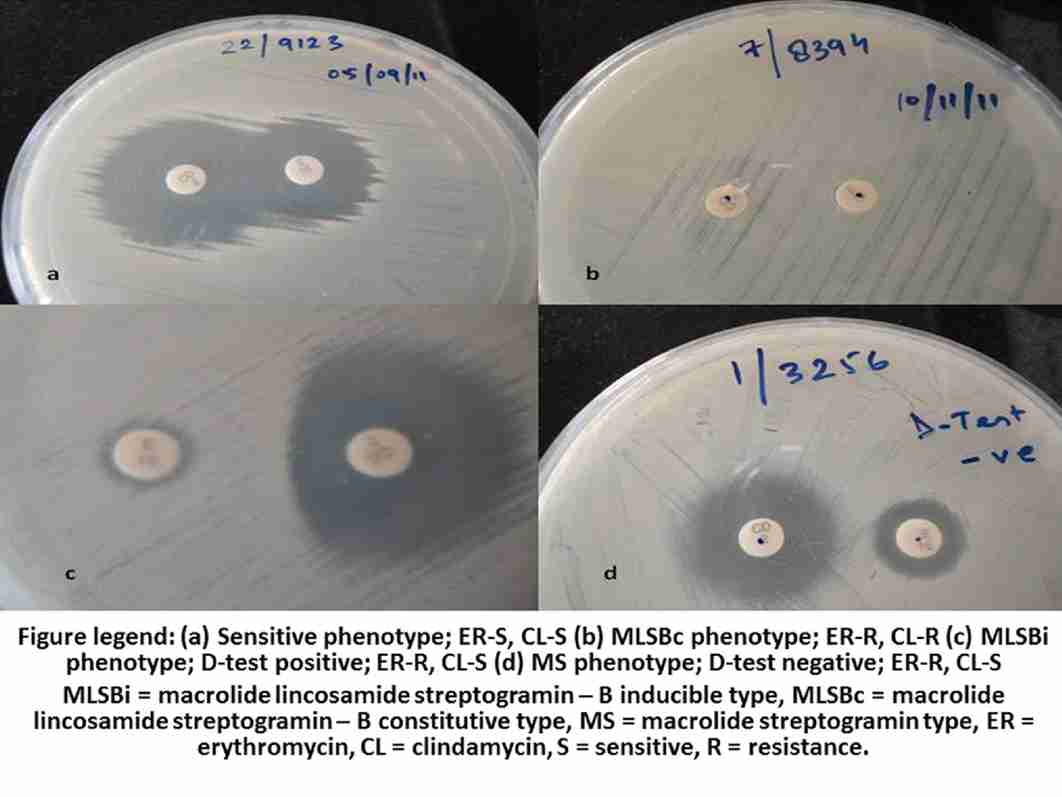
Phenotypic characterization of isolates
| Isolate | Phenotype |
| ER-R, CL-R (MLSBC phenotype), No. Positive (%) | ER-S, CL-S (Sensitive) No. Positive (%) | ER-R, CL-S (MLSBI phenotype), D-test Positive No. Positive (%) | ER-R, CL-S (MS pheno type), D-test Negative No. Positive (%) |
| MRSA (n=31) | 9 (29) | 4 (12.9) | 6 (19.4) | 12 (38.7) |
| MSSA (n=208) | 26 (12.5) | 118 (56.7) | 13 (6.3) | 51 (24.5) |
| MRCONS (n=38) | 11 (29) | 10 (26.3) | 6 (15.7) | 11 (29) |
| MSCONS (n=96) | 1 (1) | 60 (62.6) | 20 (20.8) | 15 (15.6) |
| Staphylococci (n=373) | 47 (12.6) | 192 (51.5) | 45 (12.1) | 89 (23.8) |
MRSA = methicillin resistant S.aureus, MSSA = methicillin sensitive S.aureus, MRCONS = methicillin resistant coagulase negative staphylococci, MSCONS = methicillin sensitive coagulase negative staphylococci ER = erythromycin, CL = clindamycin, R = resistant, S = sensitive
DISCUSSION
Clindamycin,a lincosamide, has long been an option for treating Staphylococcal skin, soft tissue and bone infections because of its proven efficacy, low cost, the availability of its oral and parenteral forms, tolerability,excellent tissue penetration, its good accumulation in abscesses and because no renal dosing adjustments are required. It also directly inhibits the Staphylococcal toxin production and is a useful alternative for patients who are allergic to penicillin [17]. Its good oral absorption makes it an important option in the therapy of the outpatients or as a follow up after an intravenous (IV) therapy (de-escalation). This permits an early transition to the outpatient management of the susceptible infections without the complications of a continued IV access [18]. It is effective against both the methicillin resistant and the methicillin sensitive Staphylococcal infections [1]. The increased frequency of the Staphylococcal infections, along with the changing drug susceptibility patterns, have led to a renewed interest in the CL usage [19], but the possibility of an inducible resistance to CL remains a major concern and this could limit the use of this drug [4]. To report the CL susceptibility accurately, the Staphylococci which are isolated from the clinical specimens should first be subjected to the D-test, to exclude the isolates with an induced CL resistance (MLSBi); as such isolates, when treated with CL, can undergo a rapid in vitro conversion to a constitutive resistance (MLSBc) and this may result in the CL treatment failure. Many cases of clindamycin therapy failures due to the MLSBi phenotype, have been reported in the past [9–12].
There have been various reports on the pattern of the MLSB resistance among the Staphylococci; some reports indicate a high prevalence of the MLSBi phenotype, while the others indicate an increasing frequency of the MLSBc phenotype.
Our findings were consistent with those of the studies published by other authors. A study from Karnataka [6] reported an inducible resistance in 13.1% of the Staphylococcal isolates and another study from Korea [20] reported the same in 14.6% of the isolates. We reported the same in 12.1% of our isolates.
Mohanasoundaram from Tamil Nadu [21] demonstrated MLSBi in 28% of the MRSA, in 11% MSSA and in 17% CONS. In north India [22], it was reported to be 30% in the MRSA and 10 % in the MSSA. Similarly a study from Karnataka [14] reported the same in 35.33% of the MRSA and in 11.74% of the MSSA isolates. Van der Heijden et.al., from Brazil [23] found an inducible resistance in 11.3% of the S.aureus isolates and in 13.7% CONS. We found MLSBi in 19.4% MRSA, in 6.3% MSSA, and in 19.4% CONS.
However, a study from Chicago [24] found inducible resistance in 83% of the discordant isolates among which a majority were MSSA, whereas we observed this to be 33.6% in our study and the incidence was more among the CONS isolates. In two different studies from Karnataka, the MLSBi phenotype was seen in 63% [25] and 55.26% [14] isolates among the ER-CL discordant strains of S.aureus. The same was 23% in the current study.
Fibelkorn et al., [1] showed the inducible resistance to be 29% in S.aureus and 30% in CONS. We observed the same in 7.9% of the S.aureus and in 19.4% of the CONS isolates.
The reason for this lower incidence may be the geographical and the environmental factors which were entirely different in the different clinical set ups. Moreover, our study was done in a remote place and a majority of the population belongs to the rural areas and hence is less exposed to the antimicrobial agents.
These findings suggest that the MLSBi phenotype widely varies on the basis of the geographical location, hospital environment, patient age, clinical samples which are examined, bacterial species involved and the antibiotic susceptibility profile of the bacteria [20–25]. The frequency of MLSBi ranges from 7% - 94% [26]. The MLSB resistance is the most widespread and the clinically important mechanism of resistance among the gram positive organisms due to the production of methylases and efflux proteins. The emergence of the resistance to multiple antibiotics among the gram positive organisms has left limited options for the clinicians and an appropriate therapeutic decision is not possible without the relevant antibiotic susceptibility data. This is where the D-test becomes significant.
The pattern of macrolide resistance among the Staphylococci varies in different regions. Depending upon this, the prescription rate will not be uniform in different regions. There is no substantial data on the CL prescription in India [27]. This study emphasized the prevalence of the inducible resistance among Staphylococci from the Garhwal hills of Uttarakhand and this is first study in this region. Since our hospital is an upcoming government medical institute and as molecular laboratory facilities were unavailable, so, the molecular diagnosis of these isolates was not possible. Also, the molecular markers for the erm genes are costly and inconvenient for everyday use. Patients coming to our hospital belong to rural background and majority of them are below poverty line and hence are not able to bear the heavy expenditures. In the Indian context, with the high burden of the Staphylococcal infections, where the health associated expenditures are borne by the patients, other alternatives to vancomycin are needed. Clindamycin is a good option, but the prevalence of the inducible resistance against it should be known.
The inducible resistance can be easily missed by routine in vitro susceptibility tests, when the ER and the CL discs are placed in non adjacent positions. In view of the therapeutic implications, the ER-CL disc approximation test or the D-test was found to be a simple, auxiliary, easy to perform and a reliable method which delineates the inducible (MLSBi) and the constitutive (MLSBc) resistance. So, the D-test becomes an imperative part of the antimicrobial susceptibility tests for all the Staphylococcal isolates on a routine basis. Thus, CL can be omitted in patients with infections which are caused by the strains with the MLSBi phenotype, to avoid possible therapeutic failures. The increasing prevalence of the inducible resistance (MLSBi) as compared to that of the constitutive (MLSBc) resistance among Staphylococci and the indiscriminate use of antimicrobial agents has further deteriorated the sensitivity pattern.Further epidemiological studies are required to understand this better.
MRSA = methicillin resistant S.aureus, MSSA = methicillin sensitive S.aureus, MRCONS = methicillin resistant coagulase negative staphylococci, MSCONS = methicillin sensitive coagulase negative staphylococci *Pleural Fluid, Knee Aspirate, Semen, Vaginal discharge
MRSA = methicillin resistant S.aureus, MSSA = methicillin sensitive S.aureus, MRCONS = methicillin resistant coagulase negative staphylococci, MSCONS = methicillin sensitive coagulase negative staphylococci ER = erythromycin, CL = clindamycin, R = resistant, S = sensitive