PCR Based Molecular Detection of the Gyr-B-2 Gene from the Klebsiella Sp. Isolates from Patients who were Suffering with Pneumonia and Urinary Tract Infections (UTIs)
Md. Javed Foysal1, Md. Mahbubur Rahman2, Md. Shamsul Haq Prodhan3
1 Research Assistant, USDA-Project, Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh.
2 Associate Professor, Department of Genetic Engineering and Biotechnology, Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh.
3 Associate Professor, Department of Genetic Engineering and Biotechnology, Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh.
NAME, ADDRESS, E-MAIL ID OF THE CORRESPONDING AUTHOR: Dr. Md. Shamsul Haq Prodhan, Associate Professor, Department of Genetic Engineering and Biotechnology, Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh
Phone: +8801717389379
E-mail: shamsulhp@yahoo.com
Purpose: Detection of the virulence gene is a key component in determining the pathogenicity of any isolates, because these genes act multi-functionally and multi-factorially. A gyrase specific gene primer, in combination with the PCR technology, allows the precise detection of the DNA gyrase subunit B2 gene (gyr-B-2) from different virulent microorganisms. In the present study, forward and reverse primers with lengths of 20bp and 21bp were used for the detection of the gyr-B-2 genes in the clinical isolates of the Klebsiella sp. which were collected from patients who were suffering from pneumonia and urinary tract infections (UTIs).
Materials and Methods: A total of 14 isolates viz., K1, K2, K3, K4, K5, K6, K7, K8, K9, K10, K11, K12, K13 and K14 were isolated from 3 different private medical colleges of Sylhet city.
Results: The gyr-B-2 gene which was amplified in 12 isolates viz., K1, K2, K3, K4, K5, K6, K7, K8, K10, K11, K12 and K14 gave the expected 411bp PCR product after its visualization under a gel documentation system in a 1.2% agarose gel.
Conclusions: The present study was undertaken to detect the gyrB2 gene from Klebsiella sp, which will be helpful for further scientific studies. This PCR was outstanding in the detection of the gyb-B-2 gene in pneumonia and urinary tract infections in patients, which were caused by the Klebsiella species.
Virulence gene, gyr-B-2, PCR amplification, Visualization
INTRODUCTION
The molecular techniques which have been used in recent years for the detection of pathogens and their virulent genes, have proven to be promising in the disease diagnosis and the prophylaxis. Among them, the Polymerase Chain Reaction (PCR) has been found to be the most significant one in the last couple of years [1]. The Klebsiella species are gram negative, rod shaped, non-motile bacteria that are found in the environment and also in the human intestinal tract [2,3]. Pneumonia is the commonest infection among the patients in the intensive care facilities across the world. It ranks worst among the patient morbidity and mortality cases in hospital acquired infections [4]. K. pneumoniae is the most common species which is isolated from hospital patients. In most of the patients, a variety of sites are colonized, mostly the urinary tract, with or without a serious infection [5]. Rarely can K. pneumoniae cause severe pneumonia in susceptible individuals. The clinical features of a bacteraemic infection with K. pneumoniae are due to the virulence factors which are expressed by the organism [3]. Bajaj et al. (1999) reported that the Klebsiella species caused urinary tract infections in a maximum number of cases (124, 37.35%), followed by Escherichia coli (114, 34.4%), Pseudomonas aeruginosa 32 (9.64%) and Staphylococcus aureus 23 (6.93%) [6]. Several genes are responsible for the virulence properties of the Klebsiella species, but the DNA gyrase subunit B2 gene (gyr-B-2) is the principle one that has been reported by many researchers [7]. DNA gyrase is a prokaryotic type II topoisomerase which cuts both strands of the DNA helix simultaneously in order to manage the DNA tangles and the supercoils and it is a major target of the quinolone antibacterials [7, 8]. A majority of the mutations which confer resistance to the quinolones arise within the quinolone resistance-determining region of gyrA, close to the active site (Tyr122), where DNA is bound and cleaved [7]. However, some quinolone resistance mutations are known to exist in gyrB. Liu et al. (2008) detected Klebsiella pneumoniae by PCR in infants by using the16S-23S internal transcribed spacer [1]. Catherine Dauga (2002) amplified the gyrB gene and performed a molecular phylogenetic analysis in the Enterobacteriaceae, which included Klebsiella pneumonia and Klebsiella terrigena [6].
MATERIALS AND METHODS
Collection of the Bacterial Isolates
The Klebsiella sp. bacterial isolates were collected from three different medical colleges and hospitals of the Sylhet district, Bangladesh viz. the M. A. G. Osmani Medical College Hospital, the Ibn Sina Hospital and the Ragib-Rabeya Medical College and Hospital, Bangladesh. The isolates were collected from patients who were suffering from pneumonia and urinary tract infections.
The Culture Conditions
The bacterial isolates were streaked on nutrient agar plates from a previous plate and they were incubated at 37°C overnight for the appropriate colony formation. After the incubation, a single colony of each plate was selected for re-isolation to get a pure culture in a new nutrient agar plate.
DNA Extraction
A total of 13 bacterial isolates were inoculated into nutrient broth and they were incubated overnight at 37°C and at 120rpm in a shaker incubator. The bacterial genomic DNA was extracted by using a commercial ATP Genomic DNA Extraction Kit and the extracted DNA was preserved at -20°C in an ultra freezer [Table/Fig-1].
Klebsiella sp. isolates with their isolation history
| Isolates | Collected from | Type of patient |
| K1 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | UTI |
| K2 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | Pneumonia |
| K3 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | UTI |
| K4 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | UTI |
| K5 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | Pneumonia |
| K6 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | UTI |
| K7 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | UTI |
| K8 M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | UTI |
| K9 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | UTI |
| K10 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | UTI |
| K11 | Ibn Sina Hospital, Sylhet, Bangladesh | UTI |
| K12 | Ragib-Rabeya Medical College and Hospital, Sylhet, Bangladesh | Pneumonia |
| K13 | Ibn Sina Hospital, Sylhet, Bangladesh | UTI |
| K14 | M. A. G. Osmani Medical College and Hospital, Sylhet, Bangladesh | Pneumonia |
The PCR Reaction Mixture Set Up
The PCR was performed in 25μl reaction mixtures which contained 1.2μl of the DNA template (genomic DNA of the bacteria) , 1μl of 25 mM MgCl2, 5μl of the 5x colourless reaction buffer, 0.5 μl concentration of each deoxynucleotide Triphosphate (dNTP), 1.2μl of each forward primer and reverse primer and 0.15μl of DNA polymerase along with its amplification buffer. The amplifications were carried out in a MultiGene gradient thermal cycler (Labnet International Inc. USA) [Table/Fig-2].
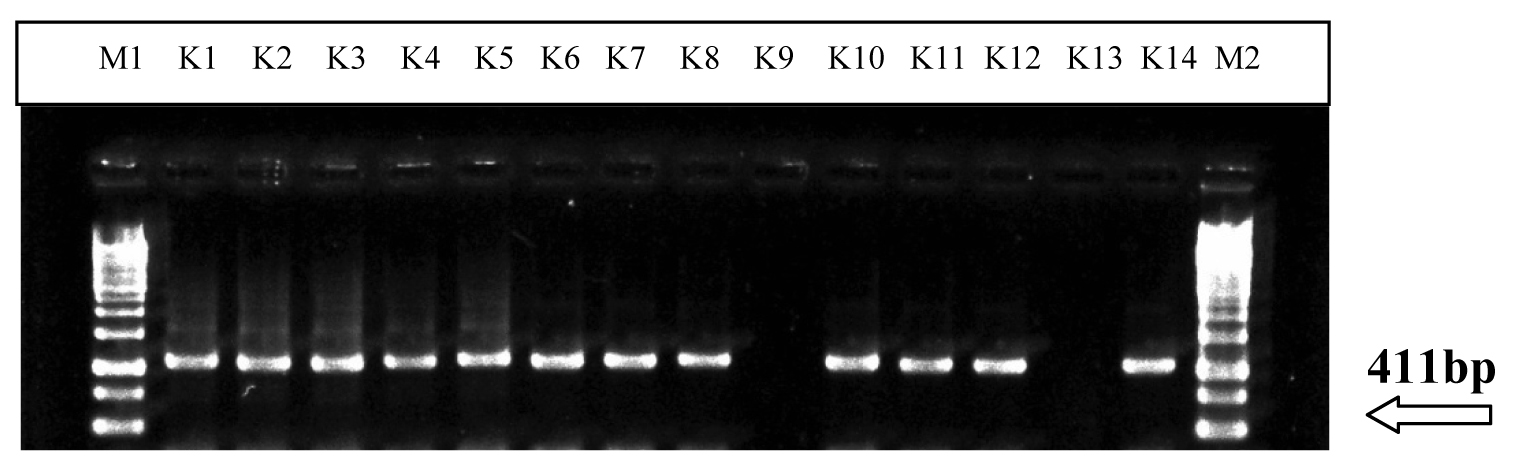
PCR amplification of gyr-B-2 gene in Klebsiella sp. with expected product length of 411bp in 1.2% agarose gel.
The Amplification Conditions
The PCR reaction was optimized with the following parameters: An initial denaturation step at 94°C for 4 min; a denaturation step at 94°C for 1 minute, annealing at 62°C for 1 minute, extension at 72°C for 90 s; and a final extension step at 72°C for 10 minutes. 35 serial cycles of reactions were performed.
RESULTS
The amplified PCR products were detected by the agarose gel electrophoresis of each amplification mixture in 1.2% agarose gels in 1% Tris-acetate-EDTA buffer. The gels were stained with an ethidium bromide solution (10mg/ml) for 20 minutes. The position of each band on the gels were then visualized and they were documented in a gel documentation system. Among the 13 isolates of Klebsiella sp., the gyr-B-2 gene was amplified in 12 isolates viz., K1, K2, K3, K4, K5, K6, K7, K8, K10, K11, K12 and K14. The sizes of the amplification products which were obtained by PCR were identical to those which were predicted from the target gyr-B-2 primers [Table/Fig-3].
Primer used for present study
| Primer Sequence (5’ to 3’) PCR product length Pathogen |
| Gyr-B-2F TCCGGCGGTCTGCACGGCGT 411bp Klebsiella sp. |
| Gyr-B-2R TTGTCCGGGTTGTACTCGTC |
DISCUSSION
The Klebsiella sp. has been reported as a common pathogen for human associated pneumonia and other diseases. Earlier studies had reported that the gyrase subunit B2 gene contributed the major virulence properties of many bacterial species and it had been used as a molecular tool for the identification of the bacterial species [8] and for the phylogenetic analysis [9–11]. It was reported that the pathogenesis of the Klebsiella sp. was multifactorial, that the mechanisms were not clearly understood and that the site of attachment and the penetration were not known [2]. In this study, we examined whether the gyr-B-2 gene was amplified in the Klebsiella sp. and whether it had a direct role in the Klebsiella sp. associated diseases. Podschun and Ullmann reported the contribution of the gyr-B-2 gene in the progression of the Klebsiella spp. and other Enterobactericeae associated diseases [2]. The gyrB gene is a single-copy gene which is present in all the bacteria which encode the ATPase domain of DNA gyrase, which is an enzyme which is essential for DNA replication [12]. In the present study, 12 isolates were found to be positive for the gyr-B-2 gene, thus revealing the distribution and the virulence properties of this gene in the Klebsiella species. This study confirmed the detection of the gyr-B-2 gene product of 411bp by using PCR as a taxonomic marker.
[1]. Liu Y, Liu C, Zheng W, Zhang X, Yu J, Gao Q, PCR detection of Klebsiella pneumoniae in infant formula based on 16S-23S internal transcribed spacerIntl J of Food Microbiol 2008 125(3):230-35. [Google Scholar]
[2]. Podschun R, Ullmann U, Klebsiella spp. as Nosocomial Pathogens: Epidemiology, Taxonomy, Typing Methods, and Pathogenicity FactorsClinical Microbiol Reviews 1998 11:589-603. [Google Scholar]
[3]. Yu VL, Hansen DS, Ko WC, Sagnimeni A, Klugman KP, Gottberg AV, Goossens Virulence Characteristics of Klebsiella and Clinical Manifestations of K. pneumonia Bloodstream InfectionsEmerging Infectious Dis. 2007 13:986-93. [Google Scholar]
[4]. George P, Sequiera A, Antimicrobial sensitivity pattern among organisms which were isolated from the endotracheal aspirates of patients with ventilator associated PneumoniaJ of Clinical and Diagnostic Res. 2011 4:3397-401. [Google Scholar]
[5]. Dhingra KR, A Case of Complicated Urinary Tract Infection: Klebsiella pneumonia Emphysematous Cystitis Presenting as Abdominal Pain in the Emergency DepartmentWest J Emerg Med. 2008 9(3):171-73. [Google Scholar]
[6]. Dauga C, Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studiesIntl J of Systematic and Evolutionary Microbiol 2002 52:531-47. [Google Scholar]
[7]. Nitiss JL, DNA Topoisomerase II and Its Growing Repertoire of Biological FunctionsNature publishing group 2009 16:1-6. [Google Scholar]
[8]. Bajaj JK, Karyakarte RP, Kulkarni JD, Deshmukh AB, Changing aetiology of urinary tract infections and emergence of drug resistance as a major problemJournal of Common Dis. 1999 31(3):181-84. [Google Scholar]
[9]. Coenye T, LiPuma JJ, Use of the gyrB gene for the identification of Pandoraea speciesFEMS Microbiol Letters 2002 208:15-19. [Google Scholar]
[10]. Yina H, Caoa L, Xiea M, Chena Q, Qiua G, Zhouc J, Bacterial diversity based on 16S rRNA and gyrB genes at Yinshan mine, ChinaSystematic and Applied Microbiol 2008 31:302-11. [Google Scholar]
[11]. Yamamoto S, Harayamapcr S, Amplification and Direct Sequencing of gyrB Genes with Universal Primers and Their Application to the Detection and Taxonomic Analysis of Pseudomonas putida StrainsApplied and Environmental Microbiol. 1995 61(3):1104-09. [Google Scholar]
[12]. Huang WM, Bacterial diversity based on type II DNA topoisomerase genesAnnu Rev Genet 1996 30:79-107. [Google Scholar]